
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
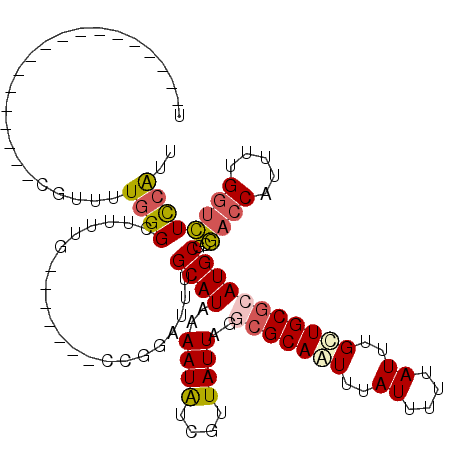
| Location | 356,836 – 356,931 |
| Length | 95 |
| Max. P | 0.925246 |

| Location | 356,836 – 356,931 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -17.11 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
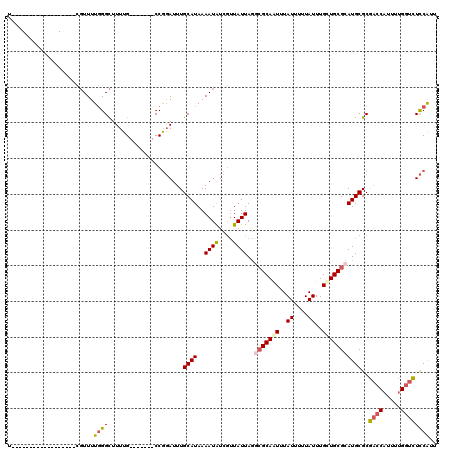
>3L_DroMel_CAF1 356836 95 + 23771897 U------------------CGUUUUGGGCUUUUG-------CCGGAUUUGCAUAAAAUAUCGUUAUUAGGCGCAGUUUAUUUUUAUUUGCUGCGCAUGCGCGACCAUUUUGGUCUCCAUU .------------------.......(((....)-------))(((...((((..((((....))))..(((((((..((....))..)))))))))))..((((.....)))))))... ( -30.00) >DroSec_CAF1 11011 113 + 1 UCGACAGUAAUUUAGAUUUCGUUUUGGGCUUUUG-------CCGGAUUUGCAUAAAAUAUCGUUAUUAGGCGCAGUUUAUUUUUAUUUGCUGCGCAUGCGCGACCAUUUUGGUCUCCAUU ..........................(((....)-------))(((...((((..((((....))))..(((((((..((....))..)))))))))))..((((.....)))))))... ( -30.00) >DroSim_CAF1 11646 95 + 1 U------------------CGUUUUGGGCUUUUG-------CCGGAUUUGCAUAAAAUAUCGUUAUUAGGCGCAGUUUAUUUUUAUUUGCUGCGCAUGCGCGACCAUUUUGGUCUCCAUU .------------------.......(((....)-------))(((...((((..((((....))))..(((((((..((....))..)))))))))))..((((.....)))))))... ( -30.00) >DroEre_CAF1 12344 113 + 1 UCAACUGUGUUUUAGAUUUCGUUUUGGGCUUUUG-------CCGGAUUUGCAUAAAAUAUCGUUAUUAGGCGCAAUUUAUUUUUAUUUGCUGCGCAUGCGCGACCAUUUCGGUCUCCAGU ..(((.((((((((.....((..((.(((....)-------)).))..))..)))))))).)))....((.((((...........)))).(((....)))((((.....)))).))... ( -25.90) >DroWil_CAF1 10229 102 + 1 U------------------UUUUUUUGGUUUUUGGUGGGGCCCAUAUUUGCAUAAAAUGUCACUAUUAAAAGCAAUUUAUUUUUAUUUGUUGCGCAUGCGCAGCCAUUUUGAAUUUCGUC .------------------........((((((((((((((....((((.....))))))).)))))))))))...............((((((....))))))................ ( -20.20) >DroYak_CAF1 12374 113 + 1 UCAACUGUAUUUUAGAUUUCGUUUUGGGCUUUUG-------CCGGAUUUGCAUAAAAUAUCGUUAUUAGCCGCAAUUUAUUUUUAUUUGCUGCGCAUGCGCGACCAUUUUGGUCUCCACU ..(((.((((((((.....((..((.(((....)-------)).))..))..)))))))).)))....((.((((...........)))).))((....))((((.....))))...... ( -25.90) >consensus U__________________CGUUUUGGGCUUUUG_______CCGGAUUUGCAUAAAAUAUCGUUAUUAGGCGCAAUUUAUUUUUAUUUGCUGCGCAUGCGCGACCAUUUUGGUCUCCAUU ........................((((.....................((((..((((....))))..(((((((..((....))..)))))))))))..((((.....)))))))).. (-17.11 = -17.58 + 0.47)

| Location | 356,836 – 356,931 |
|---|---|
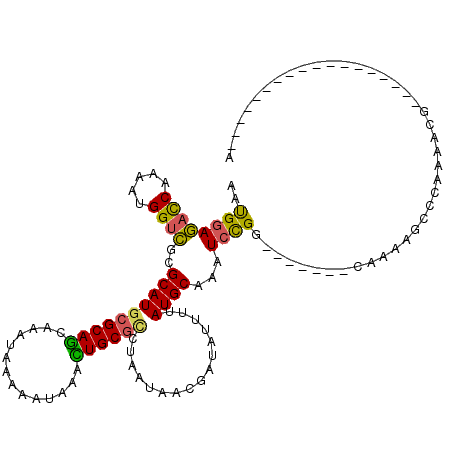
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -15.44 |
| Energy contribution | -14.91 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
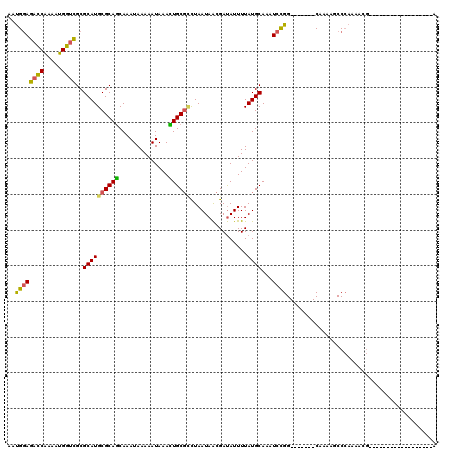
>3L_DroMel_CAF1 356836 95 - 23771897 AAUGGAGACCAAAAUGGUCGCGCAUGCGCAGCAAAUAAAAAUAAACUGCGCCUAAUAACGAUAUUUUAUGCAAAUCCGG-------CAAAAGCCCAAAACG------------------A ...(((((((.....))))..((((((((((..............))))))..((((....))))..))))...)))((-------(....))).......------------------. ( -23.74) >DroSec_CAF1 11011 113 - 1 AAUGGAGACCAAAAUGGUCGCGCAUGCGCAGCAAAUAAAAAUAAACUGCGCCUAAUAACGAUAUUUUAUGCAAAUCCGG-------CAAAAGCCCAAAACGAAAUCUAAAUUACUGUCGA ...(((((((.....))))..((((((((((..............))))))..((((....))))..))))...)))((-------(....))).......................... ( -23.74) >DroSim_CAF1 11646 95 - 1 AAUGGAGACCAAAAUGGUCGCGCAUGCGCAGCAAAUAAAAAUAAACUGCGCCUAAUAACGAUAUUUUAUGCAAAUCCGG-------CAAAAGCCCAAAACG------------------A ...(((((((.....))))..((((((((((..............))))))..((((....))))..))))...)))((-------(....))).......------------------. ( -23.74) >DroEre_CAF1 12344 113 - 1 ACUGGAGACCGAAAUGGUCGCGCAUGCGCAGCAAAUAAAAAUAAAUUGCGCCUAAUAACGAUAUUUUAUGCAAAUCCGG-------CAAAAGCCCAAAACGAAAUCUAAAACACAGUUGA ...(((((((.....))))..((((((((((..............))))))..((((....))))..))))...)))((-------(....))).......................... ( -20.54) >DroWil_CAF1 10229 102 - 1 GACGAAAUUCAAAAUGGCUGCGCAUGCGCAACAAAUAAAAAUAAAUUGCUUUUAAUAGUGACAUUUUAUGCAAAUAUGGGCCCCACCAAAAACCAAAAAAA------------------A ..........(((((((.((((....)))).).............(..((......))..))))))).........(((......))).............------------------. ( -13.60) >DroYak_CAF1 12374 113 - 1 AGUGGAGACCAAAAUGGUCGCGCAUGCGCAGCAAAUAAAAAUAAAUUGCGGCUAAUAACGAUAUUUUAUGCAAAUCCGG-------CAAAAGCCCAAAACGAAAUCUAAAAUACAGUUGA ...(((((((.....))))(((....)))................(((((...((((....))))...))))).)))((-------(....))).......................... ( -22.10) >consensus AAUGGAGACCAAAAUGGUCGCGCAUGCGCAGCAAAUAAAAAUAAACUGCGCCUAAUAACGAUAUUUUAUGCAAAUCCGG_______CAAAAGCCCAAAACG__________________A ..((((((((.....))))..((((((((((..............))))))................))))...)))).......................................... (-15.44 = -14.91 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:27 2006