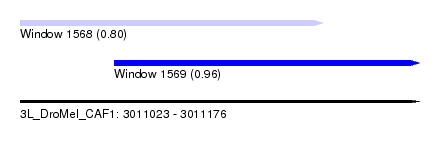
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,011,023 – 3,011,176 |
| Length | 153 |
| Max. P | 0.963504 |

| Location | 3,011,023 – 3,011,139 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -26.19 |
| Energy contribution | -29.80 |
| Covariance contribution | 3.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3011023 116 + 23771897 ACCAGUUUUAAUUUGCAUUCCUUUCGCCUCGAAG----GAGCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAAAGGUGAUGCAGGUGGCU ...(((....((((((((((((((.((.((....----)))).(((((.((((((.....))))))))))).((((((.(((.........).))))))))))))).))))))))).))) ( -41.70) >DroSec_CAF1 45311 116 + 1 ACCAGUUUUAAUUUGCAUUCCUUUCGCCUCGAAG----GAGCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAACGGUGAUGCAGGUGGGU (((.......(((((((((((....((.((....----)))).(((((.((((((.....))))))))))).((((((.(((.........).))))))))...)).))))))))).))) ( -39.80) >DroSim_CAF1 45899 116 + 1 ACCAGUUUUAAUUUGCAUUCCUUUCGCCUCGAAG----GAGCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAAAGGUGAUGCAGGUGGUU ..........((((((((((((((.((.((....----)))).(((((.((((((.....))))))))))).((((((.(((.........).))))))))))))).))))))))).... ( -41.40) >DroYak_CAF1 46615 114 + 1 AGCAAUUUUAAUUUGCAUUCCUUUCGCCUCGAAG----GAGCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUCAACUCACUUAAUCAGCAAACAGACAUGGGUGAUGCAGGUGG-- .((((.......))))........(((((((..(----((((.(((((.((((((.....))))))))))).)))))...(((((((.((........))..))))))))).))))).-- ( -39.90) >DroAna_CAF1 42364 97 + 1 ACCACUUUUAAUUUGCAUUCCUUC---UUUGUAGGACUGAGCUGCGGGAUGUCGAACUGCAGGACACCUGCCGCUGUUACUCAUUUAAUCAGGAGACAGG-------------------- ..................((((..---.....))))((((((.(((((.((((.........))))))))).)))....(((..........))).))).-------------------- ( -23.10) >consensus ACCAGUUUUAAUUUGCAUUCCUUUCGCCUCGAAG____GAGCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAAAGGUGAUGCAGGUGG_U ..........((((((((..((((.((((.(.((....((((.(((((.((((((.....))))))))))).))))...))).........(....))))).))))..)))))))).... (-26.19 = -29.80 + 3.61)


| Location | 3,011,059 – 3,011,176 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -21.18 |
| Energy contribution | -24.38 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3011059 117 + 23771897 GCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAAAGGUGAUGCAGGUGGCUGGCUGGAGGCCA---GCAACAUGCGCUUAAAAUUAAAAAC ((((((((.((((((.....))))))))))).(((((((.((((((.....((......))..))))))))))))).((((((.....))))---))....)))................ ( -45.30) >DroSec_CAF1 45347 117 + 1 GCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAACGGUGAUGCAGGUGGGUGGCUGGAGGCUA---GCAACAUGCGCUUAAAAUUAAAAAC ((.(((((.((((((.....))))))))))).((.(((((((((((.((((.(.....(....)).))))..)))))))).(((((...)))---)).))).)))).............. ( -41.30) >DroSim_CAF1 45935 117 + 1 GCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAAAGGUGAUGCAGGUGGUUGGCUGGAGGCCA---GCAACAUGCGCUUAAAAUUAAAAAC ((((((((.((((((.....))))))))))).(((((((.((((((.....((......))..))))))))))))).((((((.....))))---))....)))................ ( -42.90) >DroYak_CAF1 46651 116 + 1 GCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUCAACUCACUUAAUCAGCAAACAGACAUGGGUGAUGCAGGUGG----CUGGAGAGCAGCGGCGACAUGCGCUUAAAAUUAAAAAU ...(((((.((((((.....)))))))))))(((......(((((((.((........))..))))))).(((.((.(----(((........)))).)).))))))............. ( -36.80) >DroAna_CAF1 42401 88 + 1 GCUGCGGGAUGUCGAACUGCAGGACACCUGCCGCUGUUACUCAUUUAAUCAGGAGACAGG-----------------------------AUG---GUAACAUGCGCUUGAAAUUAAAAAC ((.(((((.((((.........))))))))).((((((((.(((((..((....))..))-----------------------------)))---)))))).)))).............. ( -25.50) >consensus GCAGCGGGAUGUCCUAAUGCAGGACACCUGCAGCUUGUACUCACUUAAUCAGCAGACAGGCAAAGGUGAUGCAGGUGG_UGGCUGGAGGCCA___GCAACAUGCGCUUAAAAUUAAAAAC ((.(((((.((((((.....))))))))))).(((((((.((((((..((........))...)))))))))))))...................((.....)))).............. (-21.18 = -24.38 + 3.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:54 2006