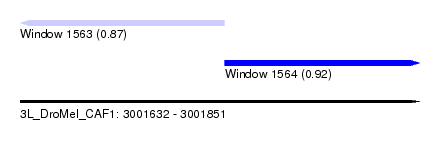
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,001,632 – 3,001,851 |
| Length | 219 |
| Max. P | 0.917902 |

| Location | 3,001,632 – 3,001,744 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.85 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -15.76 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3001632 112 - 23771897 AUUCUCGCUCCGGCUACGGCU----CUUUGGCUUCAGCUUUG-GCUUUGGAGCCGGACAAGUGUCGGAUUUGGGGGGCCGAUGGAAAGGCGCUCCUUCAAAGCGACAGCGUUAUCAA ....((((((((((..(((((----((..((((........)-)))..))))))).......))))))...((((.(((........))).))))......))))............ ( -44.80) >DroSec_CAF1 35893 116 - 1 AUUCUGGCUCCGGCUUCGGUUUUGGCUUUGGCUUCAGCUUUG-GCUUUGGAGCUGGACAAGUGUCGGAUUUGGGGAGCAGCUUAAAAGGCGCUCCUUCAAGGCGACAGCGUUAUCAA ..((..(((((((((..((((..(((....)))..))))..)-)))..)))))..))....(((((..((((((((((.(((.....)))))))).))))).))))).......... ( -47.10) >DroSim_CAF1 36495 110 - 1 AUUCUGGCUCCGGCAUCG------GCUUUGGCUUCAGCUUUG-GCUUUGGAGCCGGACAAGUGUCGGAUUUGUGGGGCAGCUUAAAAGGCGCUCCUUCAAGGCGACAGCGUUAUCAA ..(((((((((((....(------(((..(((....)))..)-))))))))))))))....(((((..((((.(((((.(((.....))))))))..)))).))))).......... ( -45.80) >DroEre_CAF1 38522 97 - 1 AUUCUGGCUCCGG------------------CUUUGGCUUUG-GCUUUGGAGCCGGACAAGUGUCGGAUUUGG-GGGCCGCUUGAAAGGCGCUCCUUCAAGGCGACCGCGUUAUCAA ..(((((((((((------------------((........)-)))..)))))))))...((((((..(((((-(((.((((.....))))..)))))))).))).)))........ ( -39.20) >DroYak_CAF1 36344 109 - 1 AUUCUGGCUCCGGCUCCAGCU------ACGGCUUUGGCUCCA-GCUUUGCAGCUGGACAAGUGUCGGAUUUGG-GGGCCGCUUGAAAGACGCUUCUCCAAGGCGACAGCGUUAUCAA .....((((((.(((..(((.------...)))..)))((((-(((....)))))))(((((.....))))))-)))))...(((..((((((((.(....).)).)))))).))). ( -41.60) >DroAna_CAF1 33756 88 - 1 ---------------------------GAGGAUUUUGCAUUGGUCUUUGGAGCCGGACAAGUGUCGGAUUUGG--GACGGCUGAAAAGAACCUGCUCGAAGGAGGCAACGUUGUCAA ---------------------------((.(((.((((...(((((((..(((((..(((((.....))))).--..)))))..)))).)))..(((....))))))).))).)).. ( -32.80) >consensus AUUCUGGCUCCGGCU_CG_________UUGGCUUCAGCUUUG_GCUUUGGAGCCGGACAAGUGUCGGAUUUGG_GGGCCGCUUAAAAGGCGCUCCUUCAAGGCGACAGCGUUAUCAA ....................................(((....((((((((((((.((....))))).......((((..((....))..)))))))))))))...)))........ (-15.76 = -16.27 + 0.51)


| Location | 3,001,744 – 3,001,851 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -29.54 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3001744 107 + 23771897 CCCGGAUUGCGCCGCACCUUGACAUAAAUUCAUUAAAUCUCUUGUCCCUGCGGCACUUGCGCCGCGGGACAGUGAGCAUUUAACGGAUUUCCUUGGCUCUUUGGACG .(((((..(.(((((.....).)..((((((.((((((((((((((((.(((((......)))))))))))).))).)))))).))))))....))).))))))... ( -38.20) >DroSec_CAF1 36009 107 + 1 CCCGGAUUGCGCCGCACCUUGACAUAAAUUCAUUAAAUCUCUUGUCCCUGCGGCACUUGCGCCCCGGGACAGCGAGCAUUUAACGGAUUUCCCUGGCUCUUUGGACG .(((((..(.(((((.....).)..((((((.((((((((((((((((.(.(((......)))).))))))).))).)))))).))))))....))).))))))... ( -32.90) >DroSim_CAF1 36605 107 + 1 CCCGGAUUGCGCCGCACCUUGACAUAAAUUCAUUAAAUCUCUUGUCCCUGCGGCAACUGCGCCCCGGGACAGCGAGCAUUUAACGGAUUUCCCUGGCUCUUUGGACG .(((((..(.(((((.....).)..((((((.((((((((((((((((.(.(((......)))).))))))).))).)))))).))))))....))).))))))... ( -32.90) >DroEre_CAF1 38619 107 + 1 CCCGGAUUGUGCCGCACCUUGACAUAAAUUCAUUAAAUCUCUGUUCCCUGCGGCACUUUCGCCCCGGGACAGCGAGCAUUUAACGGAUUUCGCUGGCUCUUUGGACG (((((...((((((((....((((.................))))...)))))))).......))))).(((((((.(((.....))))))))))............ ( -32.93) >DroYak_CAF1 36453 107 + 1 CCCGGAUAGCGCCGCACCUUGGCAUAAAUUCAUUAAAUCUCUUAUCCCUGCGGCACUUUCGCCCCGGGACAGCGAGCAUUUAACGGAUUUCGCUGGCUCUUUGGACG .(((((.(((((((.....))))..((((((.(((((((((((.((((.(.(((......)))).)))).)).))).)))))).)))))).....))).)))))... ( -34.20) >consensus CCCGGAUUGCGCCGCACCUUGACAUAAAUUCAUUAAAUCUCUUGUCCCUGCGGCACUUGCGCCCCGGGACAGCGAGCAUUUAACGGAUUUCCCUGGCUCUUUGGACG .(((((..(.(((((.....).)..((((((.((((((((((((((((.(.(((......))).)))))))).))).)))))).))))))....))).))))))... (-29.54 = -30.14 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:49 2006