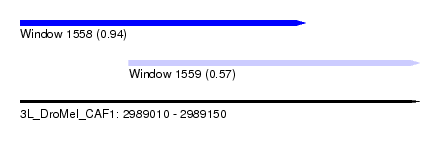
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,989,010 – 2,989,150 |
| Length | 140 |
| Max. P | 0.942555 |

| Location | 2,989,010 – 2,989,110 |
|---|---|
| Length | 100 |
| Sequences | 6 |
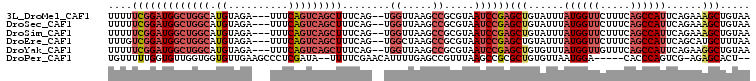
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.62 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
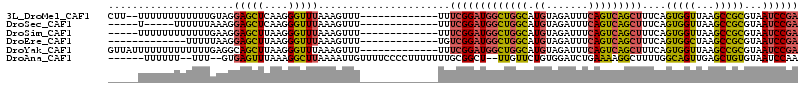
>3L_DroMel_CAF1 2989010 100 + 23771897 CUU--UUUUUUUUUUUUGUAGGAGCUCAAGGGUUUAAAGUUU-------------UUUCGGAUGGCUGGCAUGUAGAUUUCAGUCAGCUUUCAGUGGUUAAGCCGCGUAAUCCGA ...--................(((((....))))).......-------------..(((((((((((((.((.......)))))))))....((((.....))))...)))))) ( -25.60) >DroSec_CAF1 23411 92 + 1 -----U-----UUUUUUAAAGGAGCUCAAGGGUUUAAAGUUU-------------UUUCGGAUGGCUGGCAUGUAGAUUUCAGUCAGCUUUCAGUGGUUAAGCCGCGUAAUCCGA -----.-----..........(((((....))))).......-------------..(((((((((((((.((.......)))))))))....((((.....))))...)))))) ( -25.60) >DroSim_CAF1 23900 97 + 1 -----UUUUUUUUUUUUGAAGGAGCUUAAGGGUUUAAAGUUU-------------UUUCGGAUGGCUGGCAUGUAGAUUUCAGUCAGCUUUCAGUGGUUAAGCCGCGUAAUCCGA -----............((((((((((.........))))))-------------))))(((((((((((.((.......)))))))))....((((.....))))...)))).. ( -27.50) >DroEre_CAF1 25771 89 + 1 -------------UUUUUAAGGAGCUUAAGGGUUUAAAGUUU-------------UGUCGGAUGGCUGGCAUGUAGAUUUCAGUCAGCUUUCAGUGGCUAAGCCGCGUAAUCCGA -------------.......(((((((.........))))))-------------).(((((((((((((.((.......)))))))))....(((((...)))))...)))))) ( -26.00) >DroYak_CAF1 23463 102 + 1 GUUAUUUUUUUUUUUUUGAGGCAGCUUAAGGGUUUAAAGUUU-------------UUUCGGAUGGCUGGCAUGUAGAUUUCAGUCAGCUUUCAGUGGUUAAGCCGCGUAAUCCGA ...(((((....((((((((....))))))))...)))))..-------------..(((((((((((((.((.......)))))))))....((((.....))))...)))))) ( -26.00) >DroAna_CAF1 22034 103 + 1 ------UUUUUU--UUU--GUGAGUUUAAAGGCUUAAAAUUGUUUUCCCCUUUUUUUGCGGCU--UUGUUCUGUGGAUCUGAAAAGGCUUUUGGCAGUUGAGCUGUGUAAUCCAA ------......--...--.((..((....((((((..((((((....(((((((...(.((.--.......)).)....))))))).....))))))))))))....))..)). ( -20.20) >consensus _____UUUUUUUUUUUUGAAGGAGCUUAAGGGUUUAAAGUUU_____________UUUCGGAUGGCUGGCAUGUAGAUUUCAGUCAGCUUUCAGUGGUUAAGCCGCGUAAUCCGA .....................(((((....)))))......................(((((((((((((.((.......)))))))))....(((((...)))))...)))))) (-20.08 = -19.62 + -0.47)
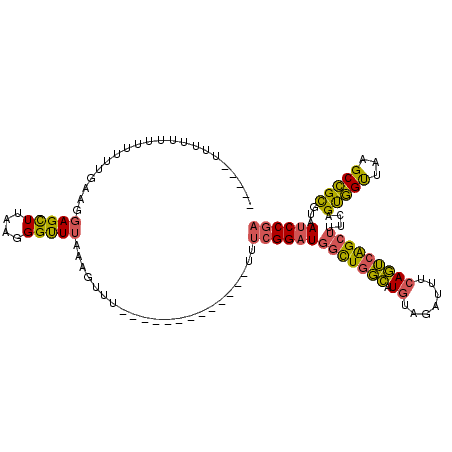
| Location | 2,989,048 – 2,989,150 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -31.20 |
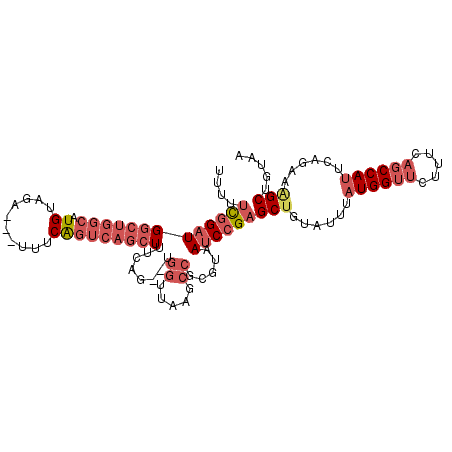
| Consensus MFE | -17.84 |
| Energy contribution | -19.95 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2989048 102 + 23771897 UUUUUCGGAUGGCUGGCAUGUAGA---UUUCAGUCAGCUUUCAG--UGGUUAAGCCGCGUAAUCCGAGCUGUAUUUAUGGUUCUUUCAGCCAUUCAGAAAGCUGUAA ....(((((((((((((.((....---...)))))))))....(--(((.....))))...))))))(((......((((((.....))))))......)))..... ( -31.90) >DroSec_CAF1 23441 102 + 1 UUUUUCGGAUGGCUGGCAUGUAGA---UUUCAGUCAGCUUUCAG--UGGUUAAGCCGCGUAAUCCGAGCUGUAUUUAUGGUUCUUUCAGCCAUUCAGAAAGCUGUAA ....(((((((((((((.((....---...)))))))))....(--(((.....))))...))))))(((......((((((.....))))))......)))..... ( -31.90) >DroSim_CAF1 23935 102 + 1 UUUUUCGGAUGGCUGGCAUGUAGA---UUUCAGUCAGCUUUCAG--UGGUUAAGCCGCGUAAUCCGAGCUGUAUUUAUGGUUCUUUCAGCCAUUCAGAAAGCUGUAA ....(((((((((((((.((....---...)))))))))....(--(((.....))))...))))))(((......((((((.....))))))......)))..... ( -31.90) >DroEre_CAF1 25798 102 + 1 UUUGUCGGAUGGCUGGCAUGUAGA---UUUCAGUCAGCUUUCAG--UGGCUAAGCCGCGUAAUCCGAGCUGUAUUUAUGGUUCUUUCAGCCAUUCAGCAUGCUUUAA ....(((((((((((((.((....---...)))))))))....(--((((...)))))...))))))((((.....((((((.....)))))).))))......... ( -34.60) >DroYak_CAF1 23503 102 + 1 UUUUUCGGAUGGCUGGCAUGUAGA---UUUCAGUCAGCUUUCAG--UGGUUAAGCCGCGUAAUCCGAGCUGUGUUUAUGGUUGUUUCAGCCAUUCAGAAGGCUGUAA ....(((((((((((((.((....---...)))))))))....(--(((.....))))...))))))(((......(((((((...)))))))......)))..... ( -33.80) >DroPer_CAF1 24713 97 + 1 UGUUUUUGGUGUUGGUGGUGUUGAAGCCCUCGAUA--UUUUCGAACAUUUUGAGCCGUUUAAGCCGCGCUGUGUUAAUGGA-----CACCCAGUCG-AGAGCACU-- (((((((((((((((..(((((((.....))))))--)..)).(((((...(.((.((....)).)).).)))))....))-----)))).....)-))))))..-- ( -23.10) >consensus UUUUUCGGAUGGCUGGCAUGUAGA___UUUCAGUCAGCUUUCAG__UGGUUAAGCCGCGUAAUCCGAGCUGUAUUUAUGGUUCUUUCAGCCAUUCAGAAAGCUGUAA ....(((((((((((((.((..........)))))))))........((.....)).....))))))(((......((((((.....))))))......)))..... (-17.84 = -19.95 + 2.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:44 2006