
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,984,119 – 2,984,233 |
| Length | 114 |
| Max. P | 0.769992 |

| Location | 2,984,119 – 2,984,233 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.35 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -10.72 |
| Energy contribution | -11.80 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2984119 114 + 23771897 GGCUUUUCAGCCGGCGAAGAGGCCCUAAAAACACUGGCACAUGCCACCAACGGAUCAAGUUUGAGGGCUUCCUCGGAAAGCCAACGAAUUAUUGUAUAGAAAAUAUUCAAAAGA ((((((((.(....)((.((((((((..((((..((((....))))((...)).....)))).)))))))).))))))))))...((((.(((........)))))))...... ( -35.80) >DroSec_CAF1 18629 89 + 1 ------------------------CG-CAAACCCAGGCACAUGCCACCAACUCAUCAAGUUUGAGGGCUUCCUCGGAAAGCCAACGAAUUAUUGUAUACCAAAUAUUCAAAAGA ------------------------.(-(((.....(((....)))............((((((..((((((....).)))))..)))))).))))................... ( -18.50) >DroSim_CAF1 18929 112 + 1 GGCUUUUCAGCCGGCGAAGAUGCCCC-UAAACUCUGGCACAUGCCACCAACGGAUCAAGUUUGAGGGCUUCCUCGGA-AGCCAACGAAUUAUUGUAUAGGAAAUAUUCAAAAGA ((((....))))(((......)))((-((.((..((((....))))...........((((((..((((((....))-))))..))))))...)).)))).............. ( -34.30) >DroEre_CAF1 20887 113 + 1 GCCUUCCUGGCCAACGAAUAGGCCUU-AAAACUCUGGCACAUGCCAACAACGGAUCAAGUUUGAGGGCUUCCUCGGAAAGCCAACGAAUUAUUGUAUGACAAAUAUUCAAAAGA .......((((...(((..(((((((-.(((((.((((....)))).(....)....))))).)))))))..)))....))))..((((..(((.....)))..))))...... ( -27.50) >DroYak_CAF1 18639 102 + 1 GCCCUCC----CAAC-------CCCA-AAAACUCUGGCACAUGCCAAGAACGGAUCAAGUUUGAGGGCUUCCUCGGAAAGCCAACGAAUUAUUGUAUGGCAAAUAUUCAAAAGA ((((((.----....-------....-.......((((....)))).((((.......))))))))))(((....))).((((((((....)))).)))).............. ( -26.90) >DroPer_CAF1 19987 86 + 1 -------------------------U-AUGGCUCACGCACAUGCCACAAACGGAUCAAGGGAGGGGGGGUGGGGGGGGGACCAAAAACGGAAUGUAUACAUUGUAUACAA--UG -------------------------.-....(((((.(...(.((.............)).)....).))))).((....))..........(((((((...))))))).--.. ( -18.82) >consensus G_CUU______C__C_______CCCU_AAAACUCUGGCACAUGCCACCAACGGAUCAAGUUUGAGGGCUUCCUCGGAAAGCCAACGAAUUAUUGUAUAGCAAAUAUUCAAAAGA ...................................(((....)))............((((((..((((((....)).))))..))))))........................ (-10.72 = -11.80 + 1.08)

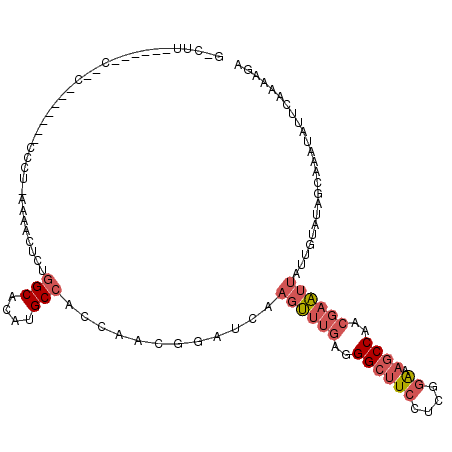
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:41 2006