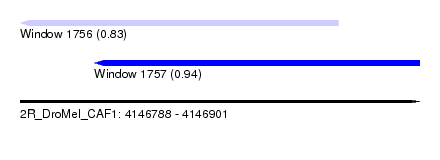
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,146,788 – 4,146,901 |
| Length | 113 |
| Max. P | 0.938480 |

| Location | 4,146,788 – 4,146,878 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.77 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4146788 90 - 20766785 AACUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAGCAACACGCAGCGAAAAUUUGCAUAAUAAUAAGUCUAUAU-----U------------ ....(((....((((((((.....)))))))).................((.....)).((((....))))..........))).....-----.------------ ( -16.70) >DroSec_CAF1 12217 90 - 1 AACUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAGCAGCACGCAGCGAAAAUUUGCAUAAUAAUAAAUCUAUAU-----C------------ ...(((.....((((((((.....)))))))))))..............((.....)).((((....))))..................-----.------------ ( -15.70) >DroSim_CAF1 16515 90 - 1 AACUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAGCAGCACGCAGCGAAAAUUUGCAUAAUAAUAAAACUAUAU-----C------------ ...(((.....((((((((.....)))))))))))..............((.....)).((((....))))..................-----.------------ ( -15.70) >DroEre_CAF1 11295 107 - 1 AACUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAGGCACACGCAGAGAAAAUUUGCAUAGUAAUGAACCUGUGAGCUAUCUAUAUUAUUUUU ...((((....((((((((.....)))))))).............((((((.....(((((.....))))).((....))..)))))).)))).............. ( -27.90) >consensus AACUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAGCAACACGCAGCGAAAAUUUGCAUAAUAAUAAAUCUAUAU_____C____________ ...(((.....((((((((.....))))))))))).....................((((.......)))).................................... (-14.23 = -14.22 + -0.00)

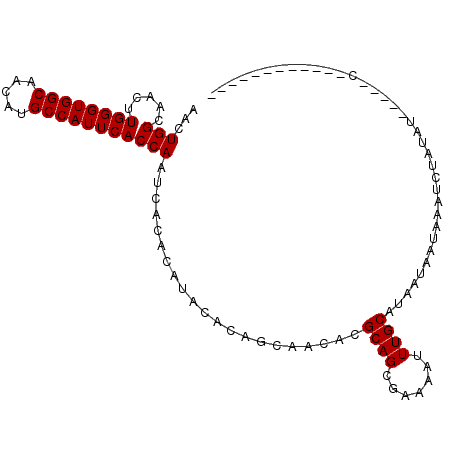
| Location | 4,146,809 – 4,146,901 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -17.67 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4146809 92 - 20766785 CGGCUAUUAAGUCUUGGC-----CAGCCAAC-----------UGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAGCAACACGCAGCGAAAAUUU .(((((........))))-----).(((...-----------.)))....((((((((.....)))))))).................((.....))........... ( -24.00) >DroPse_CAF1 5450 106 - 1 CGGCUAUUAAGUCGCGGACGAGCCAGCCAACCGAGGCACAGUUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUAUACCAGUACUCGUAGCAGCAGC-- .((((....))))((..(((((...((((((.(.....).))))))....((((((((.....)))))))).....................))))).))......-- ( -32.20) >DroGri_CAF1 10845 98 - 1 CGGCUAUUAAGUCU-GGC-----CAGCCAACCAAGACACAGUUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACACACACACACAAGGAACAG--CA--CA .(((((.......)-)))-----).((((((.........))))))....((((((((.....)))))))).............................--..--.. ( -25.30) >DroEre_CAF1 11333 92 - 1 CGGCUAUUAAGUCUUGGC-----CAGCCAAC-----------UGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAGGCACACGCAGAGAAAAUUU .(((((........))))-----).(((...-----------.(....).((((((((.....)))))))).................)))................. ( -24.40) >DroAna_CAF1 13397 85 - 1 CGGCUAUUAAGUCUUGGCCAGGCCAGCCAGU-----------UGGCAACUUGGGUGGCAACAUGGCAUUCACCAAUCACACAUACUCGUAUACCCG------------ .(((((........)))))..(((((....)-----------)))).....(((((....).(((......))).................)))).------------ ( -23.30) >DroPer_CAF1 8133 105 - 1 CGGCUAUUAAGUCGCGGCCGAGCCAGCCAACCGAGGCACAGUUGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUAUACCAGUUCUCGUAGCAGCAG--- .((((....))))((.((((((...((((((.(.....).))))))....((((((((.....)))))))).....................))))..)).))..--- ( -31.00) >consensus CGGCUAUUAAGUCUUGGC_____CAGCCAAC___________UGGCAACUUGGGUGGCAACAUGCCAUUCACCAAUCACACAUACACAAGUACACGCAGC__CA____ .((((....))))..((........((((.............))))....((((((((.....))))))))))................................... (-17.67 = -17.84 + 0.17)

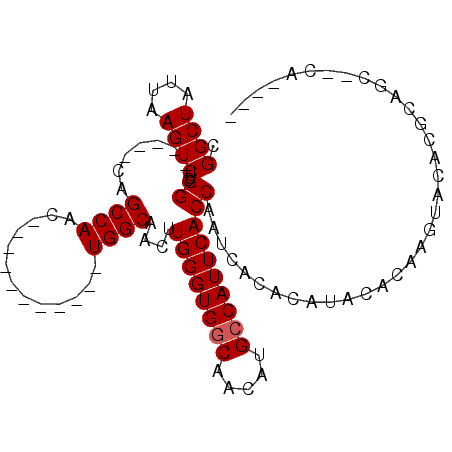
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:31 2006