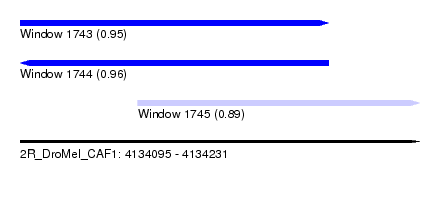
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,134,095 – 4,134,231 |
| Length | 136 |
| Max. P | 0.958266 |

| Location | 4,134,095 – 4,134,200 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -41.14 |
| Consensus MFE | -26.77 |
| Energy contribution | -28.80 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4134095 105 + 20766785 CAGCAACUCUCCUCGGACGAGGAGCAGGCCGAGCAACAUGCUCACCUGUUGCACCUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCACCGCCACAC ..............((.((.((((((((..((((.....))))...((((((....)))))---------)..((((((.---(.(....)).))---))))))))))))..)))).... ( -41.50) >DroSec_CAF1 5886 105 + 1 CAGCAACUCUCCUCGGACGAGGAGCAGGCCGAGCAACAUGCUCACCUGUUGCACCUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCGCCGCCACAC ..............((.((.((((((((..((((.....))))...((((((....)))))---------)..((((((.---(.(....)).))---))))))))))))..)))).... ( -43.10) >DroSim_CAF1 4071 105 + 1 CAGCAACUCUCCUCGGACGAGGAGCAGGCUGAGCAACAUGCUCACCUGUUGCACCUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCGCCGCCACAC ..............((.((.((((((((.(((((.....)))))..((((((....)))))---------)..((((((.---(.(....)).))---))))))))))))..)))).... ( -43.30) >DroEre_CAF1 5917 105 + 1 CAACAACUCUCCUCGGACGAGGAGCAGGUCGAGCAACAUGCUCACUUGUUGCAAUUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCACCGCCACAC ..............((.((.((((((((((((((.....))))....(((((....)))))---------)))((((((.---(.(....)).))---)))).)))))))..)))).... ( -43.10) >DroWil_CAF1 40878 114 + 1 CAACAACUCUCCUCCGAUGAGGAGCAAGCGGAACAGCAUGCUCAUCUCUUGCAUUUGCAAC------AGCGACGCGCUGGCGGUGGACAGCAACAACAAUUACCGCCACCACCGCCGCAU ........((((((....))))))...(((...((((..((((..((.((((....)))).------)).)).))))))(((((((...((.............))..)))))))))).. ( -37.52) >DroMoj_CAF1 6382 110 + 1 -AACAGCUAUCCUCGGAUGAGGAGCAGGCGGAACAGCAUGCACACCUAUUGCACUUGCAGCACCAGCAGAGACGUGUGGG---UGGAC---AGCA---AUUGCCAGCGCCGCCGCCGCAC -....((..(((((....)))))((.(((((....((.((..((((((((((....)))((....))........)))))---))..)---)((.---...))..)).))))))).)).. ( -38.30) >consensus CAACAACUCUCCUCGGACGAGGAGCAGGCCGAGCAACAUGCUCACCUGUUGCACCUGCAAC_________GACGAGCUGG___UAGCCAGCAGCA___GCUCCCUGCUCCACCGCCACAC ..............((.((.((((((((..((((....(((......(((((....)))))..............((((........)))).)))...))))))))))))..)))).... (-26.77 = -28.80 + 2.03)

| Location | 4,134,095 – 4,134,200 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -51.22 |
| Consensus MFE | -34.31 |
| Energy contribution | -34.82 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4134095 105 - 20766785 GUGUGGCGGUGGAGCAGGGAGC---UGCUGCUGGCUA---CCAGCUCGUC---------GUUGCAGGUGCAACAGGUGAGCAUGUUGCUCGGCCUGCUCCUCGUCCGAGGAGAGUUGCUG .((..(((..(((((((....)---))))(((((...---)))))))..)---------))..))...(((((((((((((.....)))).)))).((((((....)))))).))))).. ( -52.30) >DroSec_CAF1 5886 105 - 1 GUGUGGCGGCGGAGCAGGGAGC---UGCUGCUGGCUA---CCAGCUCGUC---------GUUGCAGGUGCAACAGGUGAGCAUGUUGCUCGGCCUGCUCCUCGUCCGAGGAGAGUUGCUG .((..((((((((((((....)---))))(((((...---))))))))))---------))..))...(((((((((((((.....)))).)))).((((((....)))))).))))).. ( -56.30) >DroSim_CAF1 4071 105 - 1 GUGUGGCGGCGGAGCAGGGAGC---UGCUGCUGGCUA---CCAGCUCGUC---------GUUGCAGGUGCAACAGGUGAGCAUGUUGCUCAGCCUGCUCCUCGUCCGAGGAGAGUUGCUG .((..((((((((((((....)---))))(((((...---))))))))))---------))..))...(((((((((((((.....))))).))).((((((....)))))).))))).. ( -56.70) >DroEre_CAF1 5917 105 - 1 GUGUGGCGGUGGAGCAGGGAGC---UGCUGCUGGCUA---CCAGCUCGUC---------GUUGCAAUUGCAACAAGUGAGCAUGUUGCUCGACCUGCUCCUCGUCCGAGGAGAGUUGUUG ..(((((.(.(.(((((....)---)))).)).))))---)(((((((((---------(((((....)))))....((((.....)))))))....(((((....)))))))))))... ( -47.40) >DroWil_CAF1 40878 114 - 1 AUGCGGCGGUGGUGGCGGUAAUUGUUGUUGCUGUCCACCGCCAGCGCGUCGCU------GUUGCAAAUGCAAGAGAUGAGCAUGCUGUUCCGCUUGCUCCUCAUCGGAGGAGAGUUGUUG ..((((.(((((((((((((((....))))))).))))))))(((((.(((((------.((((....)))).)).)))))..)))...))))...((((((....))))))........ ( -52.30) >DroMoj_CAF1 6382 110 - 1 GUGCGGCGGCGGCGCUGGCAAU---UGCU---GUCCA---CCCACACGUCUCUGCUGGUGCUGCAAGUGCAAUAGGUGUGCAUGCUGUUCCGCCUGCUCCUCAUCCGAGGAUAGCUGUU- ..(((((((((((((..(((..---.(((---((...---...))).))...)))..)))).(((.(..((.....))..).))).....))))...(((((....)))))..))))).- ( -42.30) >consensus GUGUGGCGGCGGAGCAGGGAGC___UGCUGCUGGCUA___CCAGCUCGUC_________GUUGCAAGUGCAACAGGUGAGCAUGUUGCUCGGCCUGCUCCUCGUCCGAGGAGAGUUGCUG ..((((((((.((((((.(.((...(((((((((......)))))..............(((((....))))).)))).)).).)))))).)))..((((((....)))))).))))).. (-34.31 = -34.82 + 0.51)
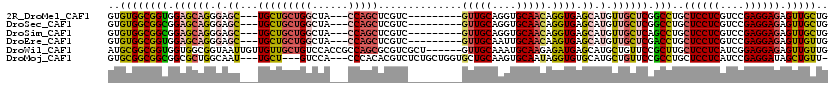
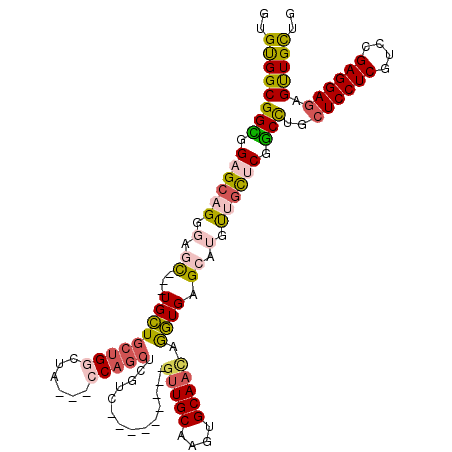
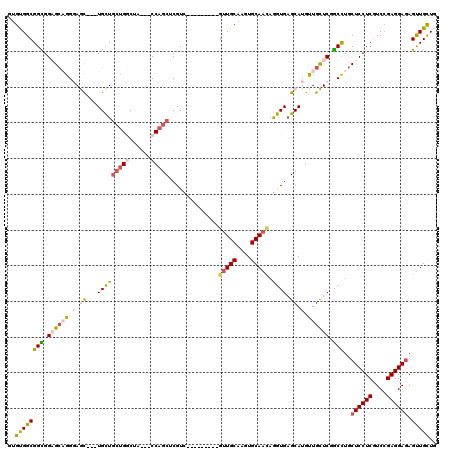
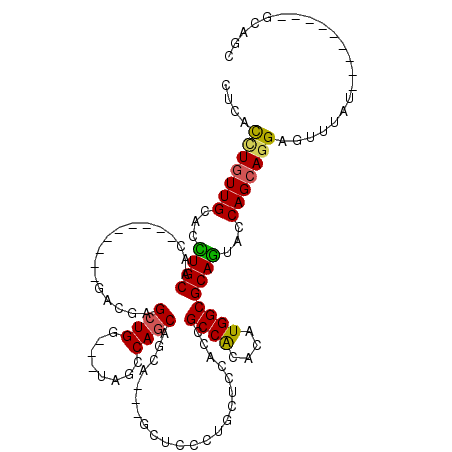
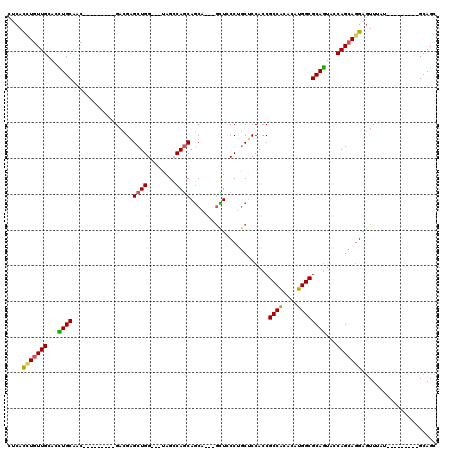
| Location | 4,134,135 – 4,134,231 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -36.79 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.03 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4134135 96 + 20766785 CUCACCUGUUGCACCUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCACCGCCACACAUGGCGCAGUACCAGCAGGAGUUUAU---------GCAGC ......((((((....)))))---------)....(((((---...))))).(((---(((((.((((..(((((((....)))))..))...)))))))))...)---------))... ( -34.50) >DroGri_CAF1 6440 114 + 1 CACACCUGUUGCAUCUGCAGCAUCAGCAGCGACGUGCUGG---UGGACAACAGCA---GUUGCCAGCGCCACCGCCACACAUGGCGCAGUAUCAGCAGGAGUUGUUGGCGCAACAGCAGC ....(((((((...((((.......((.(((((.(((((.---.......)))))---)))))..))......((((....))))))))...))))))).(((((((......))))))) ( -47.20) >DroSim_CAF1 4111 96 + 1 CUCACCUGUUGCACCUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCGCCGCCACACAUGGCGCAGUACCAGCAGGAGUUUAU---------GCAGC ....(((((((...((((...---------(.((.(((((---...)))))((((---(....))))).)).)((((....))))))))...))))))).......---------..... ( -35.20) >DroEre_CAF1 5957 96 + 1 CUCACUUGUUGCAAUUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCACCGCCACACAUGGCGCAGUACCAGCAGGAGUUUAU---------GCAGC .....(((((((....)))))---------))...(((((---...))))).(((---(((((.((((..(((((((....)))))..))...)))))))))...)---------))... ( -35.50) >DroWil_CAF1 40918 105 + 1 CUCAUCUCUUGCAUUUGCAAC------AGCGACGCGCUGGCGGUGGACAGCAACAACAAUUACCGCCACCACCGCCGCAUAUGGCGCAAUAUCAGCAGGAAUUUAU---------GCAGC ......((((((..((((...------.)))).(((((((((((((...((.............))..))))))))......))))).......))))))......---------..... ( -34.72) >DroYak_CAF1 5967 96 + 1 CUCACUUGUUGCACCUGCAAC---------GACGAGCUGG---UAGCCAGCAGCA---GCUCCCUGCUCCACCGCCGCACAUGGCGCAGUACCAGCACGAGUUUAU---------GCAGC (((..(((((((....)))))---------))...(((((---((....(.((((---(....))))).)..(((((....)))))...)))))))..))).....---------..... ( -33.60) >consensus CUCACCUGUUGCACCUGCAAC_________GACGAGCUGG___UAGCCAGCAGCA___GCUCCCUGCUCCACCGCCACACAUGGCGCAGUACCAGCAGGAGUUUAU_________GCAGC ....(((((((...((((.................((((........))))......................((((....))))))))...)))))))..................... (-25.36 = -25.03 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:19 2006