| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
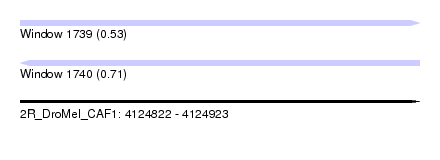
| Location | 4,124,822 – 4,124,923 |
| Length | 101 |
| Max. P | 0.711129 |

| Location | 4,124,822 – 4,124,923 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -18.00 |
| Energy contribution | -19.45 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
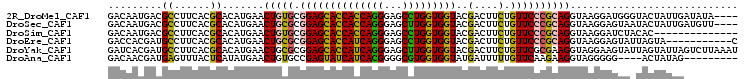
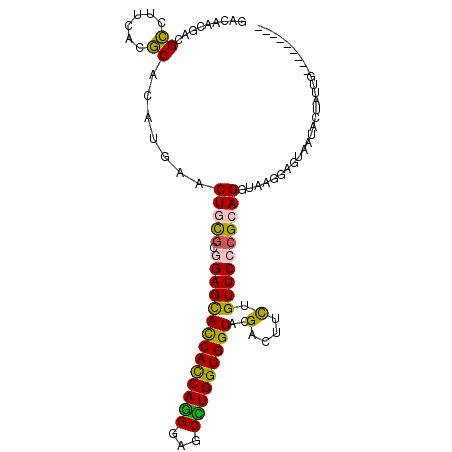
>2R_DroMel_CAF1 4124822 101 + 20766785 ----UAUAUCAAUAGUACCCAUCCUUACCUGCGGGAACAGAAGUCGUACCACCAGGCUCCCUGGUGGUGCUCCGCACAGUUCAUGUGCGUGAAGGCGUCAUUGUC ----.....((((.((.(((............))).)).((.((((((((((((((...)))))))))))((((((((.....)))))).)).))).)))))).. ( -39.50) >DroSec_CAF1 14462 101 + 1 ----AACAUCAAUAGUAUUACUCCUUACCUGCGGGAACAGAAGUCGUACCACCAAGCUCCCUGGUGGUGCUCCGCGCAGUUCAUGUGCGUGAAGGCGUCAUUGUC ----.....((((........((((.......))))...((.((((((((((((.......)))))))))((((((((.....)))))).)).))).)))))).. ( -32.00) >DroSim_CAF1 7875 91 + 1 --------------GUGUAGAUCCUUACCUGCGGGAACAGAAGUCGUACCACCAGGCUCCCUGGUGGUGCUCCGCACAGUUCAUGUGCGUGAAGGCGUCAUUGUC --------------..((((........))))(....).((.((((((((((((((...)))))))))))((((((((.....)))))).)).))).))...... ( -37.90) >DroEre_CAF1 6095 94 + 1 G-----------UACUAAUACUCCUUACCUGCGGGAACAGAAGUCGUACCACCAGGCUCCCUGAUGGUGCUCCGCGCAGUUCAUGUGCGUGAAGGCAUCGUGGUC .-----------.((((....((((.......))))...((.(((((((((.((((...)))).))))))((((((((.....)))))).)).))).)).)))). ( -30.80) >DroYak_CAF1 4301 105 + 1 AUUUAAGACUAAUACUAAUACUUCCUACCUUCGCGAACAGAAGUCGUACCACCAAGCUCCCUGAUGGUGCUCCGCGCAGUUCAUGUGCGUGAAGGCAUCGUGAUC ...........................(((((((........(..((((((.((.......)).))))))..)(((((.....)))))))))))).......... ( -26.20) >DroAna_CAF1 3384 92 + 1 ---------CUAUAGU----CCCCCUACCUUCUUGAACAAAAAUCAUACCACCACGCCCCGUGAUGAUACUCGGCACAGUUCAUAUGAGUAAACUCAUCGUUGUC ---------.......----.............(((((..............((((...)))).((.....)).....))))).(((((....)))))....... ( -9.90) >consensus _________CAAUAGUAAUACUCCUUACCUGCGGGAACAGAAGUCGUACCACCAGGCUCCCUGAUGGUGCUCCGCACAGUUCAUGUGCGUGAAGGCAUCAUUGUC .......................................((.(((((((((.((((...)))).))))))((((((((.....)))))).)).))).))...... (-18.00 = -19.45 + 1.45)

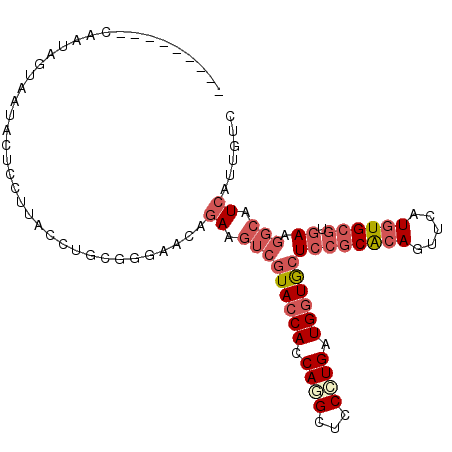
| Location | 4,124,822 – 4,124,923 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -22.75 |
| Energy contribution | -21.95 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4124822 101 - 20766785 GACAAUGACGCCUUCACGCACAUGAACUGUGCGGAGCACCACCAGGGAGCCUGGUGGUACGACUUCUGUUCCCGCAGGUAAGGAUGGGUACUAUUGAUAUA---- ..(((((..(((((((......))).(((((.((((((((((((((...)))))))))..(....).))))))))))........))))..))))).....---- ( -35.60) >DroSec_CAF1 14462 101 - 1 GACAAUGACGCCUUCACGCACAUGAACUGCGCGGAGCACCACCAGGGAGCUUGGUGGUACGACUUCUGUUCCCGCAGGUAAGGAGUAAUACUAUUGAUGUU---- ..(((((((.((((.((.(....)..(((((.((((((((((((((...)))))))))..(....).)))))))))))))))).)).....))))).....---- ( -32.50) >DroSim_CAF1 7875 91 - 1 GACAAUGACGCCUUCACGCACAUGAACUGUGCGGAGCACCACCAGGGAGCCUGGUGGUACGACUUCUGUUCCCGCAGGUAAGGAUCUACAC-------------- .........((((.(..(((((.....)))))((((((((((((((...)))))))))..(....).))))).).))))............-------------- ( -31.60) >DroEre_CAF1 6095 94 - 1 GACCACGAUGCCUUCACGCACAUGAACUGCGCGGAGCACCAUCAGGGAGCCUGGUGGUACGACUUCUGUUCCCGCAGGUAAGGAGUAUUAGUA-----------C ......((((((((.((.(....)..(((((.((((((((((((((...)))))))))..(....).)))))))))))).))).)))))....-----------. ( -31.70) >DroYak_CAF1 4301 105 - 1 GAUCACGAUGCCUUCACGCACAUGAACUGCGCGGAGCACCAUCAGGGAGCUUGGUGGUACGACUUCUGUUCGCGAAGGUAGGAAGUAUUAGUAUUAGUCUUAAAU (((...(((((((((.((((.......)))).)))).(((((((((...)))))))))...((((((....((....)).))))))....))))).)))...... ( -28.80) >DroAna_CAF1 3384 92 - 1 GACAACGAUGAGUUUACUCAUAUGAACUGUGCCGAGUAUCAUCACGGGGCGUGGUGGUAUGAUUUUUGUUCAAGAAGGUAGGGGG----ACUAUAG--------- ..((...(((((....))))).))..((.((((..(((((((((((...)))))))))))..(((((....))))))))).))..----.......--------- ( -25.10) >consensus GACAACGACGCCUUCACGCACAUGAACUGCGCGGAGCACCACCAGGGAGCCUGGUGGUACGACUUCUGUUCCCGCAGGUAAGGAGUAAUACUAUUG_________ .........((......)).......(((((.((((((((((((((...)))))))))..(....).))))))))))............................ (-22.75 = -21.95 + -0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:15 2006