
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,117,498 – 4,117,602 |
| Length | 104 |
| Max. P | 0.600827 |

| Location | 4,117,498 – 4,117,602 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
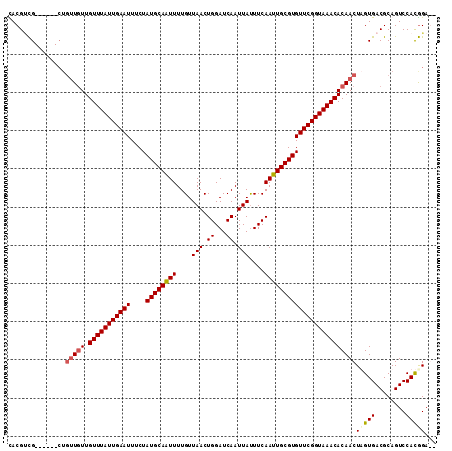
>2R_DroMel_CAF1 4117498 104 + 20766785 CACGUCG------CUGUUGUUGUUUAUUGAAUUUCUAUGCAAUUUUGUUAACUGGAUCAAUUAUUUCAAUUGCGUGUUCGGUAAACACAACUAGUGACGCAGUCCAUGGA-- ..(((((------(((((((.(((((((((((....((((((((....(((.((...)).)))....)))))))))))))))))))))))).)))))))...........-- ( -31.70) >DroSim_CAF1 30564 104 + 1 CACGUCG------CUGUUGUUGUUUAUUGAAUUUCUAUGCAAUUUUGUUAACUGGAUCAAUUAUUUCAAUUGCGUGUUCGGUAAACACAACUAGUGACGCAGUCCAUGGA-- ..(((((------(((((((.(((((((((((....((((((((....(((.((...)).)))....)))))))))))))))))))))))).)))))))...........-- ( -31.70) >DroEre_CAF1 7979 103 + 1 UACGGCG---------CUCUUGUUUAUUGAAUUUCUAUGCAGUUUUGUUAACUGGAUCAAUUACUUCAAUUGCGUGUUCGGUAAACACAACUUGUGACGCAGUCCACGGAGA .......---------((((((((((((((((....((((((((..((..............))...))))))))))))))))))))......(((........))))))). ( -21.14) >DroYak_CAF1 14521 112 + 1 UACGCUGCCUACGCUGUUGUUGUUUAUUGAAUUUCUAUGCAAUUUUGUUAACUGGAUCAAUUAUUUCAAUUGCGUGUUCGGUAAACACAACUUGUGACGCAGUCCACAGAGA ...(((((...(((.(((((.(((((((((((....((((((((....(((.((...)).)))....))))))))))))))))))))))))..)))..)))))......... ( -29.90) >consensus CACGUCG______CUGUUGUUGUUUAUUGAAUUUCUAUGCAAUUUUGUUAACUGGAUCAAUUAUUUCAAUUGCGUGUUCGGUAAACACAACUAGUGACGCAGUCCACGGA__ ...............(((((.(((((((((((....((((((((....(((.((...)).)))....))))))))))))))))))))))))(((((........)))))... (-18.85 = -19.60 + 0.75)


| Location | 4,117,498 – 4,117,602 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
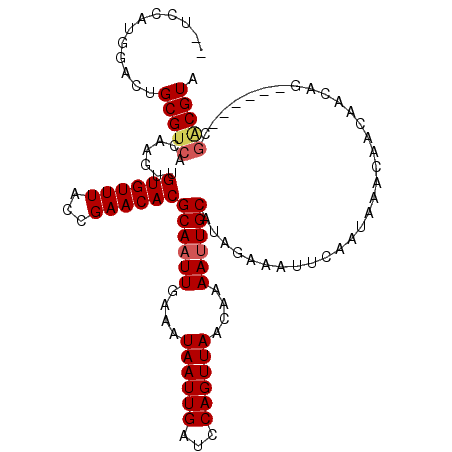
>2R_DroMel_CAF1 4117498 104 - 20766785 --UCCAUGGACUGCGUCACUAGUUGUGUUUACCGAACACGCAAUUGAAAUAAUUGAUCCAGUUAACAAAAUUGCAUAGAAAUUCAAUAAACAACAACAG------CGACGUG --..........(((((.((.((((((((((........((((((....((((((...))))))....))))))...(.....)..))))).)))))))------.))))). ( -24.20) >DroSim_CAF1 30564 104 - 1 --UCCAUGGACUGCGUCACUAGUUGUGUUUACCGAACACGCAAUUGAAAUAAUUGAUCCAGUUAACAAAAUUGCAUAGAAAUUCAAUAAACAACAACAG------CGACGUG --..........(((((.((.((((((((((........((((((....((((((...))))))....))))))...(.....)..))))).)))))))------.))))). ( -24.20) >DroEre_CAF1 7979 103 - 1 UCUCCGUGGACUGCGUCACAAGUUGUGUUUACCGAACACGCAAUUGAAGUAAUUGAUCCAGUUAACAAAACUGCAUAGAAAUUCAAUAAACAAGAG---------CGCCGUA .........((.(((((....((.((((((...)))))))).((((((.......((.(((((.....))))).)).....))))))......)).---------))).)). ( -18.00) >DroYak_CAF1 14521 112 - 1 UCUCUGUGGACUGCGUCACAAGUUGUGUUUACCGAACACGCAAUUGAAAUAAUUGAUCCAGUUAACAAAAUUGCAUAGAAAUUCAAUAAACAACAACAGCGUAGGCAGCGUA .....((...((((((.....((((((((((........((((((....((((((...))))))....))))))...(.....)..))))).))))).))))))...))... ( -23.00) >consensus __UCCAUGGACUGCGUCACAAGUUGUGUUUACCGAACACGCAAUUGAAAUAAUUGAUCCAGUUAACAAAAUUGCAUAGAAAUUCAAUAAACAACAACAG______CGACGUA ............(((((.......((((((...))))))((((((....((((((...))))))....))))))................................))))). (-16.75 = -17.38 + 0.62)

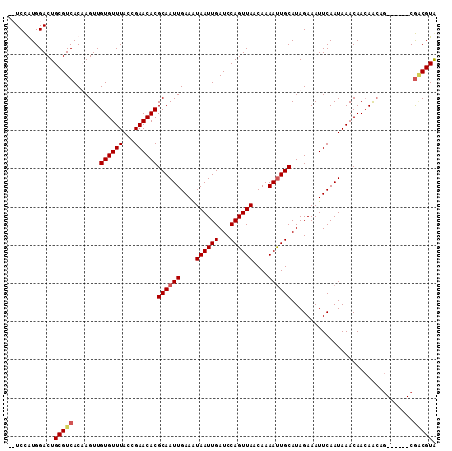
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:07 2006