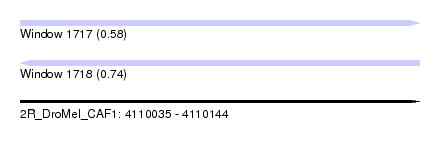
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,110,035 – 4,110,144 |
| Length | 109 |
| Max. P | 0.740831 |

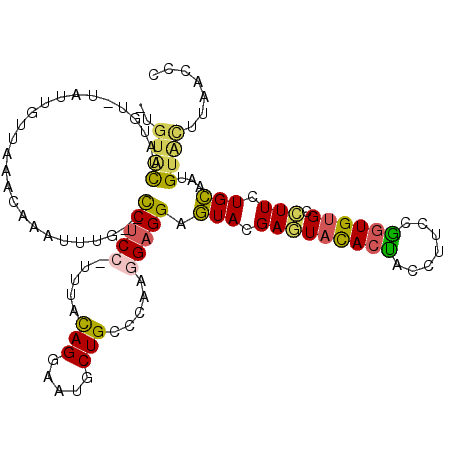
| Location | 4,110,035 – 4,110,144 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -14.23 |
| Energy contribution | -15.52 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
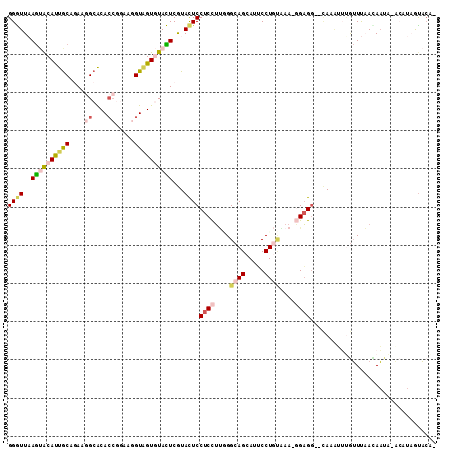
>2R_DroMel_CAF1 4110035 109 + 20766785 GGGUUAAGUACAUUGCAGAAGGCACACCGGAAGGUAGUGUACUCGUACUCCUCCUUCGGCAGCAUUCCUGUAAA-GGAGG--CAUAUUUGUUUAAGAAUAAGCAUAGUACAA .......((((..(((.((..(((((((....))).))))..))(((..(((((((..((((.....)))).))-)))))--..)))..............)))..)))).. ( -35.10) >DroGri_CAF1 13690 103 + 1 GGAUUUAGCACACCACAAAAAGCGCAAACAAAAGUUGUAUAUUCAUAUUCCUCCACAGGCAGCAUGCCUACAAA-UGUGA--UAA---UGUUUUACAAA--AGUUAAUACA- (...(((((....((((...............((..((((....))))..))....((((.....)))).....-)))).--...---((.....))..--.)))))..).- ( -11.90) >DroSec_CAF1 14107 109 + 1 GGGUUAAGUACAUUGCAGAAGGCACACCGGAAGGUAGUGUACUCGUACUCCUCCUUGGGCAGCAUUCCUGUGAA-GGAGG--CAAAUUUGUUUAACAAUAAACAUAGUACAA .......((((.((((.((..(((((((....))).))))..))......((((((.(.(((.....)))).))-)))))--)))...((((((....))))))..)))).. ( -35.10) >DroSim_CAF1 17866 106 + 1 GGGUUAAGUACAUUGCAGAAGGCACACCGGAAGGUAGUGUACUCGUACUCCUCCUUGGGCAGCAUUCCUGUGAA-GGAGG--CAAAUUUGUUUAAUAAUAUACAUAGUA--- ..((((((((..((((.((..(((((((....))).))))..))......((((((.(.(((.....)))).))-)))))--)))...)))))))).............--- ( -28.90) >DroEre_CAF1 539 108 + 1 GGGUUAAGUACAUUGCAGAAGGCACACCUGAAGGUAGUGUACUCGUACUCCUCCUUGGGCAGCAUUCCUGUAAA-GGAGG--CAUAUUCGUUUAACAAUAUACAU-GCACGU ((((..((((((((((...(((....)))....))))))))))...))))((((((..((((.....)))).))-))))(--(((.(..(((....)))..).))-)).... ( -32.50) >DroWil_CAF1 17845 108 + 1 GGAUUCAACGCAUUGCAAAACGCACACCGAAAUGUGGUAUAUUCAUACUCCUCUUGUGGCAGCAUGCCUAAUAAUUGAGGCGAAAGAAGAAUCCAUAUUC---AU-AUUCAC ((((((...((..(((.....)))..(((....(..((((....))))..).....)))..)).(((((........)))))......))))))......---..-...... ( -23.90) >consensus GGGUUAAGUACAUUGCAGAAGGCACACCGGAAGGUAGUGUACUCGUACUCCUCCUUGGGCAGCAUUCCUGUAAA_GGAGG__CAAAUUUGUUUAACAAUA_ACAUAGUACA_ ((((..((((((((((....((....)).....))))))))))...))))((((....((((.....))))....))))................................. (-14.23 = -15.52 + 1.28)

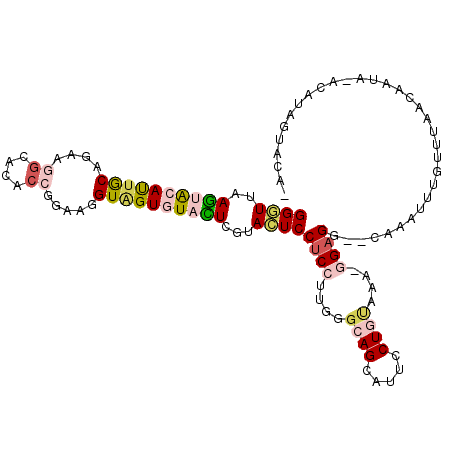
| Location | 4,110,035 – 4,110,144 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -14.89 |
| Energy contribution | -14.37 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4110035 109 - 20766785 UUGUACUAUGCUUAUUCUUAAACAAAUAUG--CCUCC-UUUACAGGAAUGCUGCCGAAGGAGGAGUACGAGUACACUACCUUCCGGUGUGCCUUCUGCAAUGUACUUAACCC ..(((((.((.(((....))).))......--(((((-(((.(((.....)))..)))))))))))))(((((((.((((....))))(((.....))).)))))))..... ( -30.10) >DroGri_CAF1 13690 103 - 1 -UGUAUUAACU--UUUGUAAAACA---UUA--UCACA-UUUGUAGGCAUGCUGCCUGUGGAGGAAUAUGAAUAUACAACUUUUGUUUGCGCUUUUUGUGGUGUGCUAAAUCC -..........--.(((((...((---(..--((...-((..(((((.....)))))..)).))..)))....))))).........(((((......)))))......... ( -18.90) >DroSec_CAF1 14107 109 - 1 UUGUACUAUGUUUAUUGUUAAACAAAUUUG--CCUCC-UUCACAGGAAUGCUGCCCAAGGAGGAGUACGAGUACACUACCUUCCGGUGUGCCUUCUGCAAUGUACUUAACCC ..(((((.((((((....))))))......--(((((-((..(((.....)))...))))))))))))(((((((.((((....))))(((.....))).)))))))..... ( -32.50) >DroSim_CAF1 17866 106 - 1 ---UACUAUGUAUAUUAUUAAACAAAUUUG--CCUCC-UUCACAGGAAUGCUGCCCAAGGAGGAGUACGAGUACACUACCUUCCGGUGUGCCUUCUGCAAUGUACUUAACCC ---......(((((((..............--(((((-((..(((.....)))...))))))).(((.(((((((((.......)))))).))).))))))))))....... ( -27.20) >DroEre_CAF1 539 108 - 1 ACGUGC-AUGUAUAUUGUUAAACGAAUAUG--CCUCC-UUUACAGGAAUGCUGCCCAAGGAGGAGUACGAGUACACUACCUUCAGGUGUGCCUUCUGCAAUGUACUUAACCC ..((..-..(((((((((....((..(((.--(((((-((..(((.....)))...))))))).))))).(((((((.......))))))).....)))))))))...)).. ( -30.50) >DroWil_CAF1 17845 108 - 1 GUGAAU-AU---GAAUAUGGAUUCUUCUUUCGCCUCAAUUAUUAGGCAUGCUGCCACAAGAGGAGUAUGAAUAUACCACAUUUCGGUGUGCGUUUUGCAAUGCGUUGAAUCC ......-..---......((((((....(((((((........))))(((((.((......)))))))))).(((((.......)))))(((((....)))))...)))))) ( -24.60) >consensus _UGUACUAUGU_UAUUGUUAAACAAAUUUG__CCUCC_UUUACAGGAAUGCUGCCCAAGGAGGAGUACGAGUACACUACCUUCCGGUGUGCCUUCUGCAAUGUACUUAACCC ..((((..........................(((((.....(((.....))).....))))).(((.(((((((((.......)))))).))).)))...))))....... (-14.89 = -14.37 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:54 2006