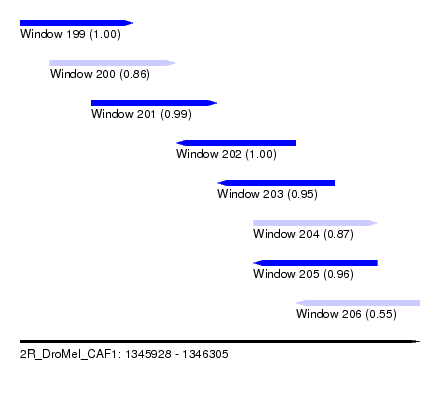
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,345,928 – 1,346,305 |
| Length | 377 |
| Max. P | 0.999013 |

| Location | 1,345,928 – 1,346,035 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.35 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -33.34 |
| Energy contribution | -33.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1345928 107 + 20766785 ------------AAACCCCAAACCAAACACGUUUGGGCCUCAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUG-GCCAUCUAUGGACAGCUUGGGUUGUCUAUGCCUGGCAGCAGC ------------(((((((((.(((((....)))))......(((......))).......))))).))))..(((.-((((..((((((((((....))))))))))..)))).))).. ( -35.70) >DroSec_CAF1 64218 108 + 1 ------------AAACCCCAAACCAAACACGUUCGGGCCCCAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUGGGCCAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGC ------------....(((.(((.......))).)))..((((((...(((.(.(........)).)))...))))))((((..((((((((((....))))))))))..))))...... ( -32.50) >DroSim_CAF1 42408 107 + 1 ------------AAACCCCAAACCAAACACGUUCGGGCCCCAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUG-GCCAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGC ------------(((((((((..((((...((.(((.............))).))..))))))))).))))..(((.-((((..((((((((((....))))))))))..)))).))).. ( -32.42) >DroEre_CAF1 45507 119 + 1 ACCCCAAGCCCCAAACCCCAAACCAAACACGUUUGGGCCUCGUGUUUUACUGCACUUUUUGUUGCGAGUUUUAUGUG-GCCAUCUAUGGACAGCUUUGGCUGUCUAUGCCUGGCGGCAGC .......((..((((.(((((((.......)))))))....((((......))))..))))..))........(((.-((((..((((((((((....))))))))))..)))).))).. ( -41.60) >DroYak_CAF1 32825 107 + 1 ------------AAACCCCAAACCAAACACGUUUGGGCCCCAUGUUUUAGUGCACUUUUUGUUGAGAGUUUUAUGUG-GCCAUCUUUGGACAGCUUUGGCUGUCUAUGCCUGGCAGCAGC ------------....(((((((.......))))))).............(((((((((....))))))........-((((....((((((((....))))))))....)))).))).. ( -33.30) >consensus ____________AAACCCCAAACCAAACACGUUUGGGCCCCAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUG_GCCAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGC ................(((((((.......)))))))............((((((((((....)))))).........((((..((((((((((....))))))))))..)))).)))). (-33.34 = -33.98 + 0.64)

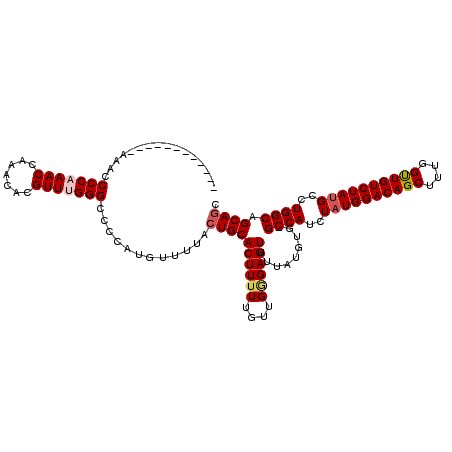
| Location | 1,345,956 – 1,346,075 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1345956 119 + 20766785 CAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUG-GCCAUCUAUGGACAGCUUGGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGCU ...........((......((.((((.((....(((.-((((..((((((((((....))))))))))..)))).))))))))).))(((((..(..........)..)))))....)). ( -33.10) >DroSec_CAF1 64246 119 + 1 CAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUGGGCCAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGC- (((((...(((.(.(........)).)))...))))).((((..((((((((((....))))))))))..))))((((((((((......))))...............))))))....- ( -32.56) >DroSim_CAF1 42436 118 + 1 CAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUG-GCCAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGC- (((((...(((.(.(........)).)))...)))))-((((..((((((((((....))))))))))..))))((((((((((......))))...............))))))....- ( -32.56) >DroEre_CAF1 45547 118 + 1 CGUGUUUUACUGCACUUUUUGUUGCGAGUUUUAUGUG-GCCAUCUAUGGACAGCUUUGGCUGUCUAUGCCUGGCGGCAGCCCCGCCACAAUGGCUCUUAUUUUAUGUUCGUUGCUUAGC- .((((......)))).....((.(((((.....(((.-((((..((((((((((....))))))))))..)))).))).....(((.....)))............))))).)).....- ( -40.10) >DroYak_CAF1 32853 116 + 1 CAUGUUUUAGUGCACUUUUUGUUGAGAGUUUUAUGUG-GCCAUCUUUGGACAGCUUUGGCUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUUUU--UUGUUCGUUGCUUAGC- ...........(((.........(((((.((((((((-((......((((((((....)))))))).....(((....)))..))))).))))).))))).--........))).....- ( -31.17) >consensus CAUGUUUUACUGCACUUUUUGUUGGGAGUUUUAUGUG_GCCAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGC_ ...........((((((((....)))))).........((((..((((((((((....))))))))))..))))((((((((((......))))...............))))))..)). (-29.38 = -29.46 + 0.08)

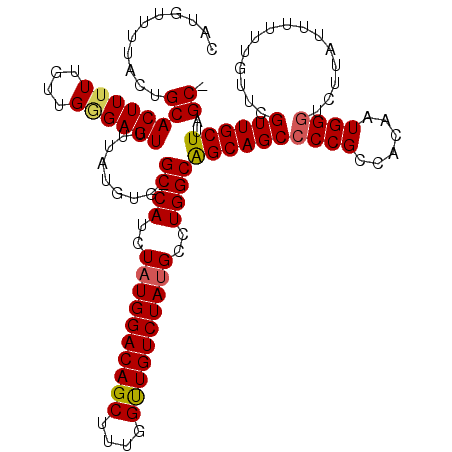
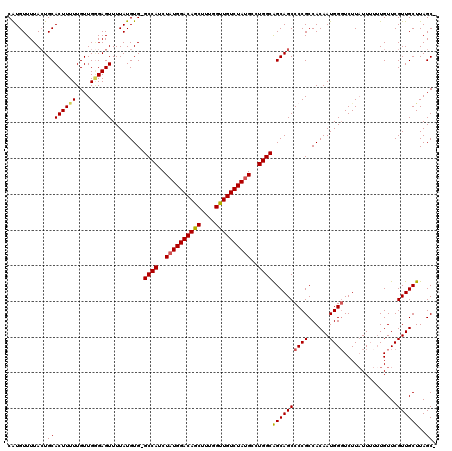
| Location | 1,345,995 – 1,346,114 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 94.63 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1345995 119 + 20766785 CAUCUAUGGACAGCUUGGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGCUGUAGCUGUAAAAAUCAUUUUAUGAGUUUGCAUAAAAUUU ....((((((((((....))))))))))...(((.(((((...((.(((((..(........)..))).)).))...))))).))).........((((((((......)))))))).. ( -30.50) >DroSec_CAF1 64286 113 + 1 CAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGC------UGUAAAAAUCAUUUUAUGAGUUUGCAUAAAAUUU .....(((((((((....)))))))))((..(((((((((((((......))))...........))).))))))..))------..........((((((((......)))))))).. ( -28.00) >DroSim_CAF1 42475 113 + 1 CAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGC------UGUAAAAAUCAUUUUAUGAGUUUGCAUAAAAUUU .....(((((((((....)))))))))((..(((((((((((((......))))...........))).))))))..))------..........((((((((......)))))))).. ( -28.00) >DroEre_CAF1 45586 113 + 1 CAUCUAUGGACAGCUUUGGCUGUCUAUGCCUGGCGGCAGCCCCGCCACAAUGGCUCUUAUUUUAUGUUCGUUGCUUAGC------UGUAAAAAUCAUUUUAUGAGUUUGCAUAAAAUUU .....(((((((((....)))))))))(((((((((.....))))))....)))....(((((((((.....((((...------.((((((....))))))))))..))))))))).. ( -34.70) >DroYak_CAF1 32892 111 + 1 CAUCUUUGGACAGCUUUGGCUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUUUU--UUGUUCGUUGCUUAGC------UGUAAAAAUCAUUUUAUGAGUUUGCAUAAAAUUU ......((((((((....)))))))).((..(((((((((((((......)))).......--..))).))))))..))------..........((((((((......)))))))).. ( -29.40) >consensus CAUCUAUGGACAGCUUUGGUUGUCUAUGCCUGGCAGCAGCCCCGCCACAAUGGGUCUUAUUUUUUGUUCGUUGCUUAGC______UGUAAAAAUCAUUUUAUGAGUUUGCAUAAAAUUU .....(((((((((....)))))))))((..(((((((((((((......))))...........))).))))))..))................((((((((......)))))))).. (-26.68 = -26.68 + -0.00)

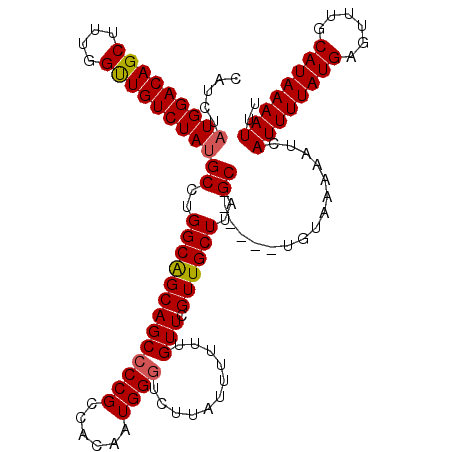
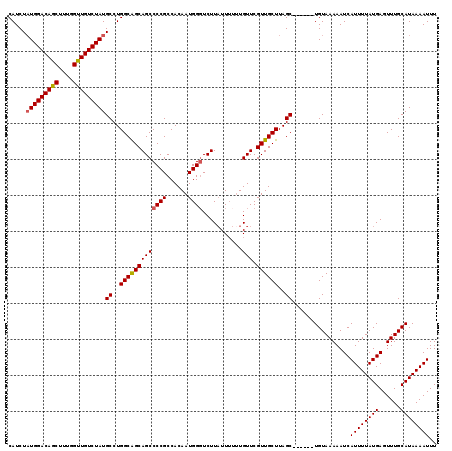
| Location | 1,346,075 – 1,346,188 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 97.09 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1346075 113 - 20766785 AAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACAAAAUUUUAUGCAAACUCAUAAAAUGAUUUUUACAGCUAC ....((((....))))((((......(((((....)))))..)))).............((((((((((.((.....(((((((......))))))))).))))))).))).. ( -25.50) >DroSec_CAF1 64365 108 - 1 AAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACAAAAUUUUAUGCAAACUCAUAAAAUGAUUUUUACA----- ....((((....))))((((......(((((....)))))..))))................(((((((.((.....(((((((......))))))))).))))))).----- ( -22.80) >DroSim_CAF1 42554 108 - 1 AAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACAAAAUUUUAUGCAAACUCAUAAAAUGAUUUUUACA----- ....((((....))))((((......(((((....)))))..))))................(((((((.((.....(((((((......))))))))).))))))).----- ( -22.80) >DroEre_CAF1 45665 108 - 1 AAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCACCGGGCACAAUAAACAUGGGCGUGAAAACCACAAAAUUUUAUGCAAACUCAUAAAAUGAUUUUUACA----- ....((((....))))((((......(((((....)))))..))))................(((((((.((.....(((((((......))))))))).))))))).----- ( -21.70) >DroYak_CAF1 32969 108 - 1 AAUUGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGAGCACAAUAAACAUGGGCGUGAAAACCACAAAAUUUUAUGCAAACUCAUAAAAUGAUUUUUACA----- ....((((....))))((((......(((((....)))))..))))................(((((((.((.....(((((((......))))))))).))))))).----- ( -23.50) >consensus AAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACAAAAUUUUAUGCAAACUCAUAAAAUGAUUUUUACA_____ ....((((....))))((((......(((((....)))))..))))................(((((((.((.....(((((((......))))))))).)))))))...... (-22.88 = -22.72 + -0.16)

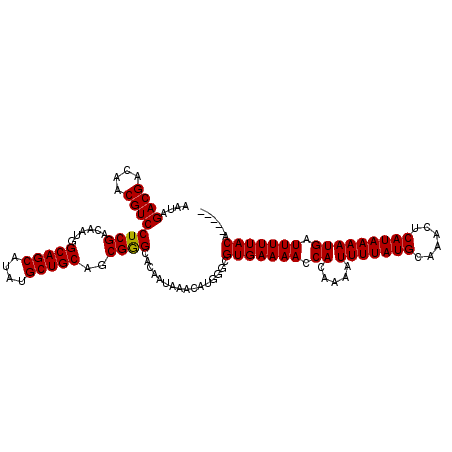
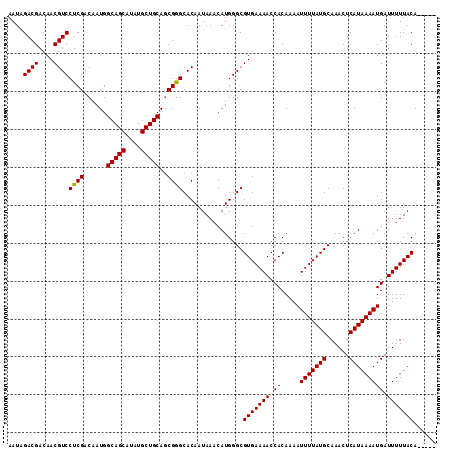
| Location | 1,346,114 – 1,346,225 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1346114 111 - 20766785 ---UCAGCUGAUGCGGCUUAGCCUGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACA ---...(((.((((((((......)))))...............((((....))))((((......(((((....)))))..)))).........))).)))(((.....))). ( -30.30) >DroSec_CAF1 64399 111 - 1 ---UCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACA ---...(((.(((.......(((((.((................((((....))))..........(((((....))))))))))))........))).)))(((.....))). ( -32.16) >DroSim_CAF1 42588 114 - 1 UCCUCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACA ......(((.(((.......(((((.((................((((....))))..........(((((....))))))))))))........))).)))(((.....))). ( -32.16) >DroEre_CAF1 45699 110 - 1 ---UCCACUGAUGCGGCUCAGCCUGGCUGGA-ACAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCACCGGGCACAAUAAACAUGGGCGUGAAAACCACA ---............(((((((((((.....-............((((....))))..........(((((....))))).))))))..........)))))(((.....))). ( -29.80) >DroYak_CAF1 33003 111 - 1 ---UCAGCUGAUACGGCUUAGCCCAGCUGGAAAUAAAAUCAAUUGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGAGCACAAUAAACAUGGGCGUGAAAACCACA ---..(((((...)))))..(((((...................((((....))))((((......(((((....)))))..))))...........)))))(((.....))). ( -31.40) >consensus ___UCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGCAGCGGGCACAAUAAACAUGGGCGUGAAAACCACA .....(((((...)))))..(((((...................((((....))))((((......(((((....)))))..))))...........)))))(((.....))). (-27.88 = -27.72 + -0.16)

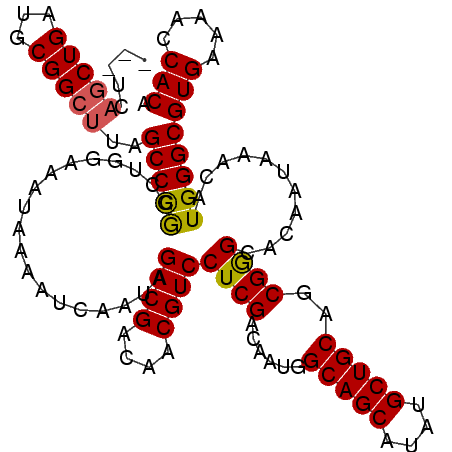
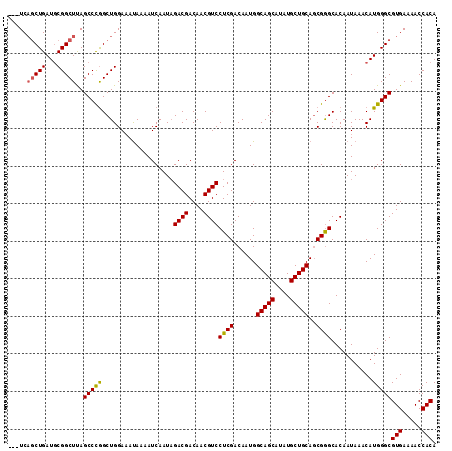
| Location | 1,346,148 – 1,346,265 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -33.48 |
| Energy contribution | -34.56 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1346148 117 + 20766785 GCAGCAUAUGCUGCCAUUGUCGAGGACGUUGUCGUCUAUUGAUUUUAUUUCCAGCCAGGCUAAGCCGCAUCAGCUGA---GCGGCUUCGCCCGAAUUCUGUGACGUCCAGGGUUCAGGGU (((((....)))))....((((((((((....))))).)))))..........(((.(((.(((((((.((....))---))))))).))).((((((((.(....)))))))))..))) ( -46.70) >DroSec_CAF1 64433 117 + 1 GCAGCAUAUGCUGCCAUUGUCGAGGACGUUGUCGUCUAUUGAUUUUAUUUCCAGCCGGGCUAAGCCGCAUCAGCUGA---GCGGCUUCGCCCGAAUUCUGUGACGUCCAGGGUUGAGGGU (((((....)))))....((((((((((....))))).)))))...(((..(((((((((.(((((((.((....))---))))))).)))).....(((.(....)))))))))..))) ( -49.20) >DroSim_CAF1 42622 120 + 1 GCAGCAUAUGCUGCCAUUGUCGAGGACGUUGUCGUCUAUUGAUUUUAUUUCCAGCCGGGCUAAGCCGCAUCAGCUGAGGAGCGGCUUCGCCCGAAUUCUGUGACGUCCAGGGUUGAGGGC (((((....)))))....((((((((((....))))).)))))......(((...(((((.(((((((..(......)..))))))).)))))(((((((.(....))))))))..))). ( -48.30) >DroEre_CAF1 45733 110 + 1 GCAGCAUAUGCUGCCAUUGUCGAGGACGUUGUCGUCUAUUGAUUUUGU-UCCAGCCAGGCUGAGCCGCAUCAGUGGA------GCUUCGCUCGAAUUCUGUGACUU---CUCCCAGGGGC (((((....)))))((..((((((((((....))))).)))))..))(-((.(((.(((((...((((....)))))------)))).))).)))(((((.((...---.)).))))).. ( -36.30) >DroYak_CAF1 33037 114 + 1 GCAGCAUAUGCUGCCAUUGUCGAGGACGUUGUCGUCAAUUGAUUUUAUUUCCAGCUGGGCUAAGCCGUAUCAGCUGA---ACGGCUUCGCACCAAUUCUGGGACUU---GGGCUAGGAAU (((((....))))).....(((((((((....))))............((((((.(((((.(((((((.((....))---))))))).)).)))...)))))))))---))......... ( -40.90) >consensus GCAGCAUAUGCUGCCAUUGUCGAGGACGUUGUCGUCUAUUGAUUUUAUUUCCAGCCGGGCUAAGCCGCAUCAGCUGA___GCGGCUUCGCCCGAAUUCUGUGACGUCCAGGGUUAAGGGU (((((....)))))....((((((((((....))))).)))))......(((((.(((((.(((((((.((....))...))))))).)))))....))).))................. (-33.48 = -34.56 + 1.08)

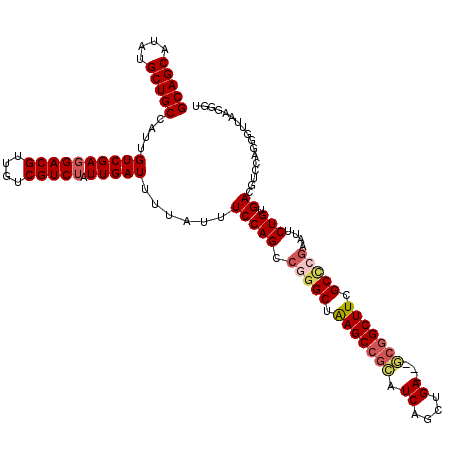
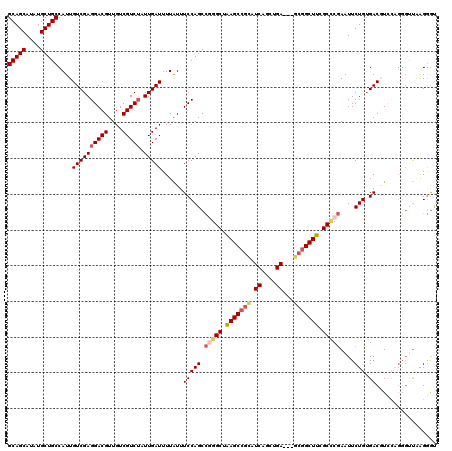
| Location | 1,346,148 – 1,346,265 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -33.42 |
| Energy contribution | -34.34 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1346148 117 - 20766785 ACCCUGAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGC---UCAGCUGAUGCGGCUUAGCCUGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGC ..((........))..(((.(((...((((((.(((((((---((....)).))))))).)))))))))...............((((....))))...)))....(((((....))))) ( -41.10) >DroSec_CAF1 64433 117 - 1 ACCCUCAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGC---UCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGC ..((........))..(((.(((...((((((.(((((((---((....)).))))))).)))))))))...............((((....))))...)))....(((((....))))) ( -43.30) >DroSim_CAF1 42622 120 - 1 GCCCUCAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGCUCCUCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGC ..((........))..(((.(((...((((((.(((((((..(......)..))))))).)))))))))...............((((....))))...)))....(((((....))))) ( -42.70) >DroEre_CAF1 45733 110 - 1 GCCCCUGGGAG---AAGUCACAGAAUUCGAGCGAAGC------UCCACUGAUGCGGCUCAGCCUGGCUGGA-ACAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGC .((....))..---..(((......(((.(((.(.((------(((.(....).))...))).).))).))-)...........((((....))))...)))....(((((....))))) ( -28.40) >DroYak_CAF1 33037 114 - 1 AUUCCUAGCCC---AAGUCCCAGAAUUGGUGCGAAGCCGU---UCAGCUGAUACGGCUUAGCCCAGCUGGAAAUAAAAUCAAUUGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGC ...........---..(((((((...(((.((.(((((((---((....)).))))))).))))).))))..............((((....))))...)))....(((((....))))) ( -37.30) >consensus ACCCUCAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGC___UCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUCAAUAGACGACAACGUCCUCGACAAUGGCAGCAUAUGCUGC ................(((.(((....(((((.(((((((...((....)).))))))).))))).)))...............((((....))))...)))....(((((....))))) (-33.42 = -34.34 + 0.92)

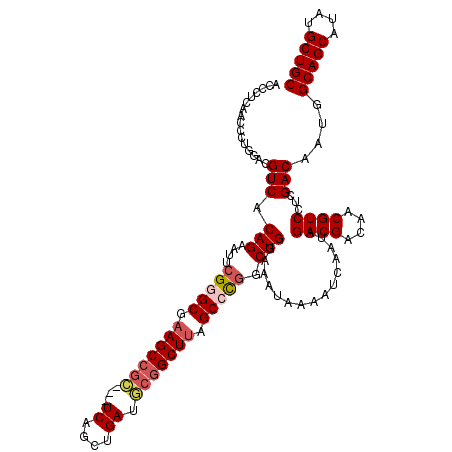
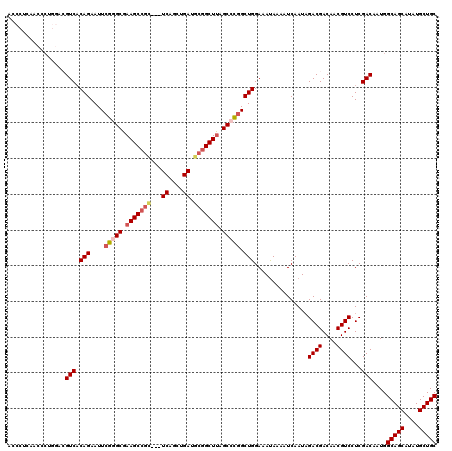
| Location | 1,346,188 – 1,346,305 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.66 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1346188 117 - 20766785 UUAGACCAGCGCGUACAUACAAGAAAUGCACAACCACUGAACCCUGAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGC---UCAGCUGAUGCGGCUUAGCCUGGCUGGAAAUAAAAUC .....(((((((((...........))))........(((..((........))...))).......(((((.(((((((---((....)).))))))).)))))))))).......... ( -36.60) >DroSec_CAF1 64473 117 - 1 UUAGACCGGCGCGUACCCACAAGAAAUGCACAACCACUGAACCCUCAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGC---UCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUC .....(((((((((...........))))........(((..((........))...))).......(((((.(((((((---((....)).))))))).)))))))))).......... ( -38.10) >DroSim_CAF1 42662 120 - 1 UUAGAGCAGCGCGUACCCACAAAAAAUGCACAACCACUGAGCCCUCAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGCUCCUCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUC ......((((((((.((..........((.((.....)).))..........)))))).........(((((.(((((((..(......)..))))))).)))))))))........... ( -36.35) >DroEre_CAF1 45773 110 - 1 UCAGACCGGCGCGUAAACACAACUCGUAUGCCAGCACUGAGCCCCUGGGAG---AAGUCACAGAAUUCGAGCGAAGC------UCCACUGAUGCGGCUCAGCCUGGCUGGA-ACAAAAUC .....((((((((((...........)))))(((..(((((((.(.(((((---...((.(.(....)..).))..)------))).)....).))))))).)))))))).-........ ( -32.60) >DroYak_CAF1 33077 114 - 1 UUAGUCCGGCGCGUACACACAAGAAAUGCACAUCCGCUGAAUUCCUAGCCC---AAGUCCCAGAAUUGGUGCGAAGCCGU---UCAGCUGAUACGGCUUAGCCCAGCUGGAAAUAAAAUC ....((((((((((...........))))..(((.(((((((..((.((((---((.((...)).)))).))..))..))---))))).)))..(((...)))..))))))......... ( -32.70) >consensus UUAGACCGGCGCGUACACACAAGAAAUGCACAACCACUGAACCCUCAACCCUGGACGUCACAGAAUUCGGGCGAAGCCGC___UCAGCUGAUGCGGCUUAGCCCGGCUGGAAAUAAAAUC .....(((((((((...........))))......................................(((((.(((((((...((....)).))))))).)))))))))).......... (-24.90 = -25.66 + 0.76)

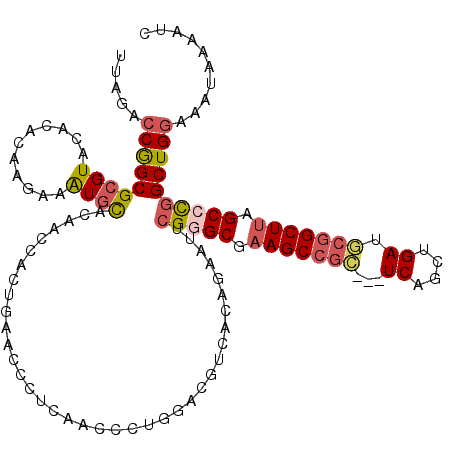
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:04 2006