
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,100,067 – 4,100,178 |
| Length | 111 |
| Max. P | 0.997104 |

| Location | 4,100,067 – 4,100,161 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -28.45 |
| Energy contribution | -27.90 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
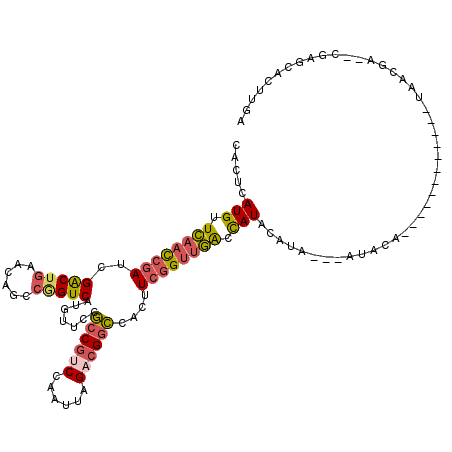
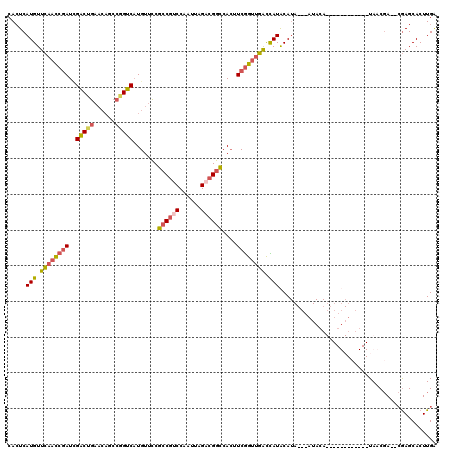
>2R_DroMel_CAF1 4100067 94 + 20766785 CACAAACGAUUUGUCCACUCAUGUUCAACCGAUAGACUGAACAGCCGGUCAUGUUCCGCCGUCCAAUUAGACGGCCACUUCGGUUGGCCAUACA- ....................(((..(((((((..(((((......))))).......((((((......))))))....)))))))..)))...- ( -30.70) >DroSec_CAF1 4161 95 + 1 CACAAACGAUUUGUCCACUCAUGUUCAACCGAUCGACUGAACAGCCAGUCAUGUUCCGCCGCCCAAUUAGACGGCCACUUCGGUUGGCCAUACAU ....................(((..(((((((..(((((......))))).......((((.(......).))))....)))))))..))).... ( -26.50) >DroSim_CAF1 4142 95 + 1 CACAAACGAUUUGUCCACUCAUGUUCAACCGAUCGGCUGAACAGCCGGUCAUGUUCCGCCGUCCAAUUAGACGGCCACUUCGGUUGACUAUACAU ....................(((.((((((((((((((....)))))))).......((((((......))))))......)))))).))).... ( -33.70) >consensus CACAAACGAUUUGUCCACUCAUGUUCAACCGAUCGACUGAACAGCCGGUCAUGUUCCGCCGUCCAAUUAGACGGCCACUUCGGUUGGCCAUACAU ....................(((.((((((((..(((((......))))).......((((((......))))))....)))))))).))).... (-28.45 = -27.90 + -0.55)

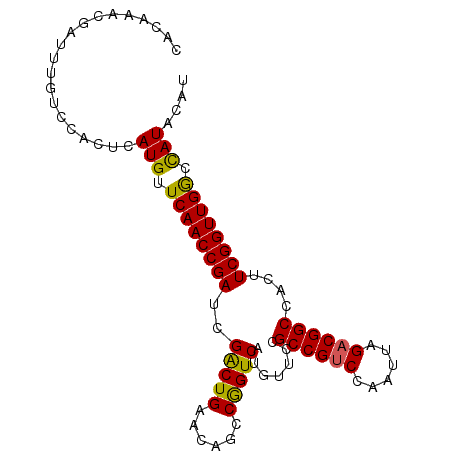
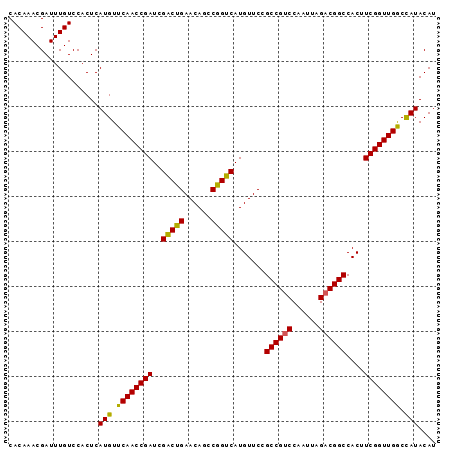
| Location | 4,100,067 – 4,100,161 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -31.84 |
| Energy contribution | -31.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
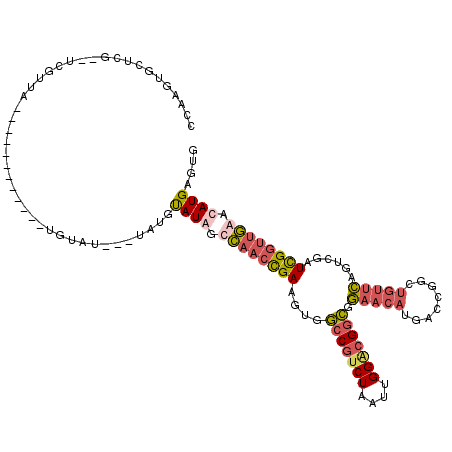
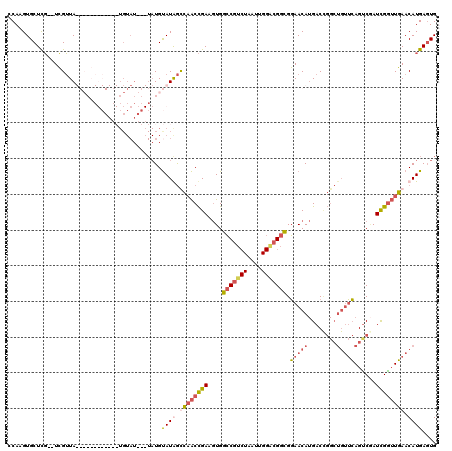
>2R_DroMel_CAF1 4100067 94 - 20766785 -UGUAUGGCCAACCGAAGUGGCCGUCUAAUUGGACGGCGGAACAUGACCGGCUGUUCAGUCUAUCGGUUGAACAUGAGUGGACAAAUCGUUUGUG -..((((..(((((((....(((((((....))))))).(((((........)))))......)))))))..)))).(..(((.....)))..). ( -33.60) >DroSec_CAF1 4161 95 - 1 AUGUAUGGCCAACCGAAGUGGCCGUCUAAUUGGGCGGCGGAACAUGACUGGCUGUUCAGUCGAUCGGUUGAACAUGAGUGGACAAAUCGUUUGUG ...((((..(((((((.((.(((((((....)))))))...)).(((((((....))))))).)))))))..)))).(..(((.....)))..). ( -36.00) >DroSim_CAF1 4142 95 - 1 AUGUAUAGUCAACCGAAGUGGCCGUCUAAUUGGACGGCGGAACAUGACCGGCUGUUCAGCCGAUCGGUUGAACAUGAGUGGACAAAUCGUUUGUG ...(((..((((((((.((.(((((((....)))))))...)).....(((((....))))).))))))))..))).(..(((.....)))..). ( -35.90) >consensus AUGUAUGGCCAACCGAAGUGGCCGUCUAAUUGGACGGCGGAACAUGACCGGCUGUUCAGUCGAUCGGUUGAACAUGAGUGGACAAAUCGUUUGUG ...((((..(((((((.((.(((((((....)))))))...))......((((....))))..)))))))..)))).(..(((.....)))..). (-31.84 = -31.73 + -0.11)

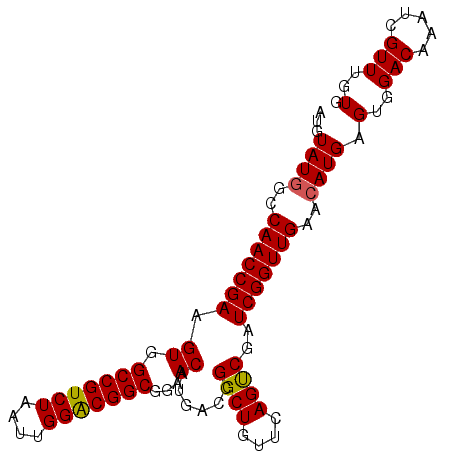
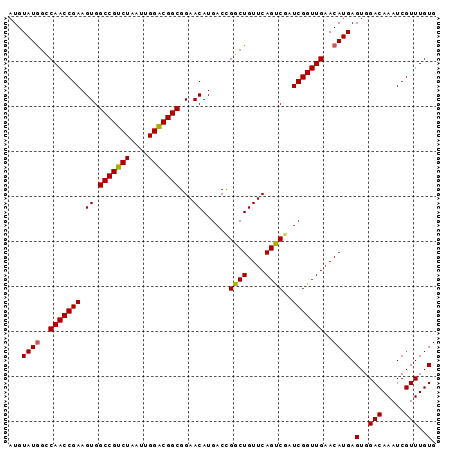
| Location | 4,100,082 – 4,100,178 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.33 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.05 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4100082 96 + 20766785 CACUCAUGUUCAACCGAUAGACUGAACAGCCGGUCAUGUUCCGCCGUCCAAUUAGACGGCCACUUCGGUUGGCCAUACA----------------------UAACGA--CGAGCACUUGA ..((((((..(((((((..(((((......))))).......((((((......))))))....)))))))..))).(.----------------------....).--.)))....... ( -31.00) >DroSec_CAF1 4176 116 + 1 CACUCAUGUUCAACCGAUCGACUGAACAGCCAGUCAUGUUCCGCCGCCCAAUUAGACGGCCACUUCGGUUGGCCAUACAUAU--AUACAUUUGUAUCUGCAUAACGA--CGAGCACUUGG ..((((((..(((((((..(((((......))))).......((((.(......).))))....)))))))..))).((.((--(((....))))).))........--.)))....... ( -30.00) >DroSim_CAF1 4157 118 + 1 CACUCAUGUUCAACCGAUCGGCUGAACAGCCGGUCAUGUUCCGCCGUCCAAUUAGACGGCCACUUCGGUUGACUAUACAUAUACAUACAUAUGUAUCUGCAUAACGA--CGAGCACUUGG ..(((....((((((((((((((....)))))))).......((((((......))))))......))))))..((((((((......))))))))...........--.)))....... ( -40.70) >DroWil_CAF1 10336 88 + 1 CAAUCAUGUUAACUCAAUUGACUCC-------GUC-UGUUGCCCCCU-----U----GCGCAGAUCAGCAUAUUAUGCAUA---AUACA------------UAUCCAGCAAAGCACUUGA .............((((.((.((..-------(((-(((.((.....-----.----))))))))..((....((((....---...))------------))....))..)))).)))) ( -11.90) >consensus CACUCAUGUUCAACCGAUCGACUGAACAGCCGGUCAUGUUCCGCCGUCCAAUUAGACGGCCACUUCGGUUGACCAUACAUA___AUACA____________UAACGA__CGAGCACUUGA .....(((.((((((((..(((((......))))).......((((((......))))))....)))))))).)))............................................ (-18.24 = -19.05 + 0.81)

| Location | 4,100,082 – 4,100,178 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.33 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -18.06 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4100082 96 - 20766785 UCAAGUGCUCG--UCGUUA----------------------UGUAUGGCCAACCGAAGUGGCCGUCUAAUUGGACGGCGGAACAUGACCGGCUGUUCAGUCUAUCGGUUGAACAUGAGUG ......(((((--((((((----------------------(((.(((....))).....(((((((....)))))))...)))))).))))((((((..(....)..)))))).)))). ( -35.40) >DroSec_CAF1 4176 116 - 1 CCAAGUGCUCG--UCGUUAUGCAGAUACAAAUGUAU--AUAUGUAUGGCCAACCGAAGUGGCCGUCUAAUUGGGCGGCGGAACAUGACUGGCUGUUCAGUCGAUCGGUUGAACAUGAGUG ......(((((--(.((((((((.((((....))))--...))))))))(((((((.((.(((((((....)))))))...)).(((((((....))))))).)))))))...)))))). ( -45.70) >DroSim_CAF1 4157 118 - 1 CCAAGUGCUCG--UCGUUAUGCAGAUACAUAUGUAUGUAUAUGUAUAGUCAACCGAAGUGGCCGUCUAAUUGGACGGCGGAACAUGACCGGCUGUUCAGCCGAUCGGUUGAACAUGAGUG ......(((((--(.((((((((.((((((....)))))).)))))).((((((((.((.(((((((....)))))))...)).....(((((....))))).)))))))))))))))). ( -48.40) >DroWil_CAF1 10336 88 - 1 UCAAGUGCUUUGCUGGAUA------------UGUAU---UAUGCAUAAUAUGCUGAUCUGCGC----A-----AGGGGGCAACA-GAC-------GGAGUCAAUUGAGUUAACAUGAUUG ((((.((((((((((..((------------(((..---...)))))......((.(((.(..----.-----.).))))).))-).)-------)))).)).))))............. ( -19.90) >consensus CCAAGUGCUCG__UCGUUA____________UGUAU___UAUGUAUAGCCAACCGAAGUGGCCGUCUAAUUGGACGGCGGAACAUGACCGGCUGUUCAGUCGAUCGGUUGAACAUGAGUG ...........................................((((.((((((((....(((((((....))))))).(((((........)))))......)))))))).)))).... (-18.06 = -19.88 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:45 2006