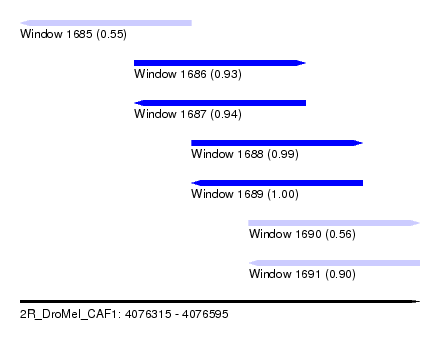
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,076,315 – 4,076,595 |
| Length | 280 |
| Max. P | 0.999867 |

| Location | 4,076,315 – 4,076,435 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -40.78 |
| Energy contribution | -40.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4076315 120 - 20766785 CCCGAGCCGGAGAAGUUCGAUCCCAGUCGCUUCGACCCGGAGGAAGUCAAGAACCGCCAUCCGAUGGCCUAUUUGCCCUUUGGCGAUGGACCAAGGAACUGCAUUGGUCUGCGGUUCGGU .((((((((.(((.((((..(((..((((...))))..))).(....)..)))).((..(((..(((((...(((((....))))).)).))).)))...)).....))).)))))))). ( -44.60) >DroSec_CAF1 59078 120 - 1 CCCGAGCCGGAAAAGUUCGAUCCCAGUCGCUUCGAUCCGGAGGAAGUCAAGAACCGCCAUCCGAUGGCCUACUUGCCCUUUGGCGAUGGACCAAGGAACUGCAUUGGACUGCGGUUCGGA .((((((((....((((((((..((((..((......((((((..((.....))..)).)))).(((((...(((((....))))).)).)))))..)))).)))))))).)))))))). ( -43.50) >DroSim_CAF1 67284 120 - 1 CCCGAGCCGGAAAAGUUCGAUCCCAGUCGCUUCGAUCCGGAGGAAGUCAAGAACCGCCAUCCGAUGGCCUACUUGCCCUUUGGCGAUGGACCAAGGAACUGCAUUGGACUGCGGUUCGGA .((((((((....((((((((..((((..((......((((((..((.....))..)).)))).(((((...(((((....))))).)).)))))..)))).)))))))).)))))))). ( -43.50) >DroEre_CAF1 60399 120 - 1 CCCGAUCCGGAAAGGUUCGAUCCCAGUCGCUUUGAUCCGGAGGAAGUCAAGAACCGCCAUCCGAUGGCCUACUUGCCCUUUGGCGAUGGACCGCGGAACUGCAUUGGACUGCGGUUCGGA .((..((((((..((..((((....))))..))..)))))).........(((((((..((((((((((...(((((....))))).)).))(((....)))))))))..))))))))). ( -47.50) >DroYak_CAF1 63363 120 - 1 CCCGAACCGGAAAAGUUCGAUCCCAGUCGCUUUGAUCCGGAGGAAGUCAAAAACCGCCAUCCGAUGGCCUACUUGCCCUUUGGCGAUGGACCUCGGAACUGCAUUGGACUGCGGUUCGGA .((((((((....((((((((..((((...........((.((..........)).)).(((((.((((...(((((....))))).)).))))))))))).)))))))).)))))))). ( -45.50) >consensus CCCGAGCCGGAAAAGUUCGAUCCCAGUCGCUUCGAUCCGGAGGAAGUCAAGAACCGCCAUCCGAUGGCCUACUUGCCCUUUGGCGAUGGACCAAGGAACUGCAUUGGACUGCGGUUCGGA .((((((((....((((((((..(((((.........((((((..((.....))..)).)))).(((((...(((((....))))).)).)))..).)))).)))))))).)))))))). (-40.78 = -40.98 + 0.20)

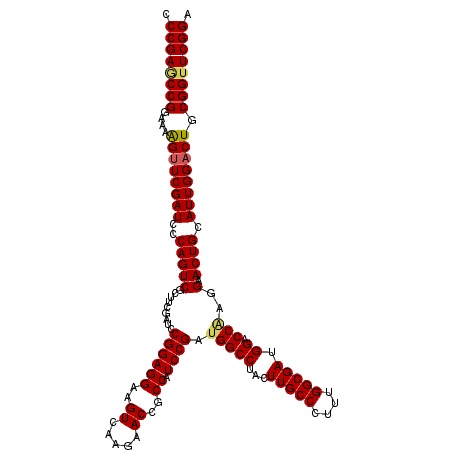
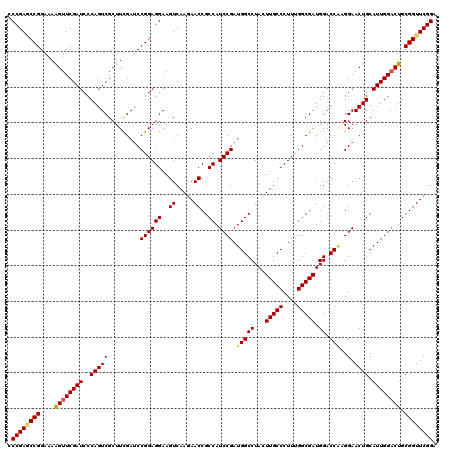
| Location | 4,076,395 – 4,076,515 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -44.36 |
| Consensus MFE | -36.40 |
| Energy contribution | -37.36 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4076395 120 + 20766785 CCGGGUCGAAGCGACUGGGAUCGAACUUCUCCGGCUCGGGAUAUAUCUCCGGAUCGUGGUGGAUAUUAUGCACCGGAAUCAGGGCAAGAAUGCCCUUGUCUAGCACAAUAUCUGAGUUGG ((.(((((...))))).))...........(((((((((.((((...(((((..((((((....))))))..)))))...((((((....))))))...........))))))))))))) ( -41.60) >DroSec_CAF1 59158 120 + 1 CCGGAUCGAAGCGACUGGGAUCGAACUUUUCCGGCUCGGGAUAUAUCUCCGGAUCGUGGUGGAUAUUGUGCACGGGUAUGAGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGG (((((((....(((((((((........)))))).)))(((......))).)))).))).(((((((((((...((.(..(((((......)))))..))).)))))))))))....... ( -48.80) >DroSim_CAF1 67364 120 + 1 CCGGAUCGAAGCGACUGGGAUCGAACUUUUCCGGCUCGGGAUAUAUCUCCGGAUCGUGGUGGAUAUUGUGCACGGGUAUGAGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGG (((((((....(((((((((........)))))).)))(((......))).)))).))).(((((((((((...((.(..(((((......)))))..))).)))))))))))....... ( -48.80) >DroEre_CAF1 60479 120 + 1 CCGGAUCAAAGCGACUGGGAUCGAACCUUUCCGGAUCGGGAUAUAUCUCAGGAUCGUGGUGAAUAUUGUGAACGGGAAUCAGGGCAAGGACGCCCUUGUCCAGCACAAUAUCCGAGUGGG .(((((((..((((((((((((((.((.....)).)))......)))))))..))))..))).(((((((....(((...(((((......)))))..)))..)))))))))))...... ( -43.70) >DroYak_CAF1 63443 120 + 1 CCGGAUCAAAGCGACUGGGAUCGAACUUUUCCGGUUCGGGAUAUAUCUCAGGGUCGUGAUGAAUAUUAUGGACGGGAAUCAGGGCAAGGACGCCCUUGUCCAGCACAAUAUCUGUGUUGG (((..(((..((((((.((((((........))))))((((....))))..))))))..)))..........)))......((((((((....))))))))((((((.....)))))).. ( -38.90) >consensus CCGGAUCGAAGCGACUGGGAUCGAACUUUUCCGGCUCGGGAUAUAUCUCCGGAUCGUGGUGGAUAUUGUGCACGGGAAUCAGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGG (((((((.....((((((((........)))))).))((((....))))..)))).))).(((((((((((...((.(..(((((......)))))..))).)))))))))))....... (-36.40 = -37.36 + 0.96)

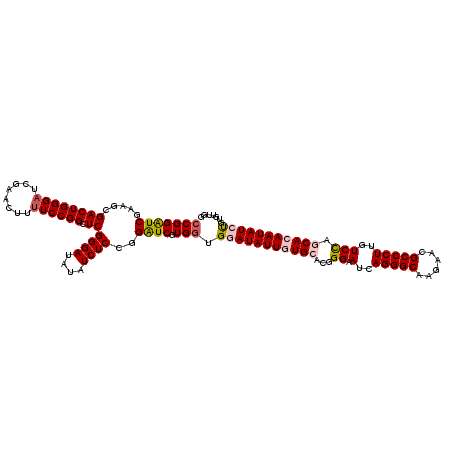
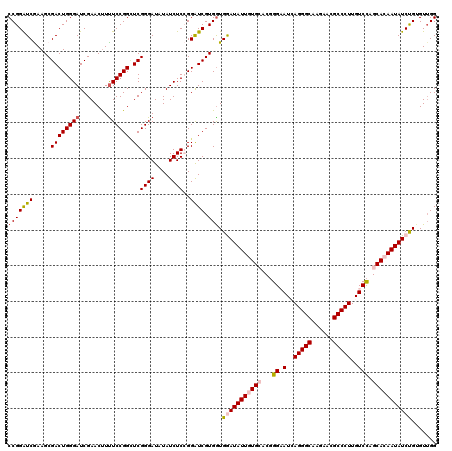
| Location | 4,076,395 – 4,076,515 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -31.84 |
| Energy contribution | -33.08 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4076395 120 - 20766785 CCAACUCAGAUAUUGUGCUAGACAAGGGCAUUCUUGCCCUGAUUCCGGUGCAUAAUAUCCACCACGAUCCGGAGAUAUAUCCCGAGCCGGAGAAGUUCGAUCCCAGUCGCUUCGACCCGG ....(((.((((((((((..((..((((((....))))))...))....))))))))))...........(((......))).)))((((.((((..((((....))))))))...)))) ( -39.20) >DroSec_CAF1 59158 120 - 1 CCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCUCAUACCCGUGCACAAUAUCCACCACGAUCCGGAGAUAUAUCCCGAGCCGGAAAAGUUCGAUCCCAGUCGCUUCGAUCCGG ........(((((((((((((...((((((....)))))).....))).))))))))))........((.(((......))).)).(((((......((((....))))......))))) ( -38.60) >DroSim_CAF1 67364 120 - 1 CCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCUCAUACCCGUGCACAAUAUCCACCACGAUCCGGAGAUAUAUCCCGAGCCGGAAAAGUUCGAUCCCAGUCGCUUCGAUCCGG ........(((((((((((((...((((((....)))))).....))).))))))))))........((.(((......))).)).(((((......((((....))))......))))) ( -38.60) >DroEre_CAF1 60479 120 - 1 CCCACUCGGAUAUUGUGCUGGACAAGGGCGUCCUUGCCCUGAUUCCCGUUCACAAUAUUCACCACGAUCCUGAGAUAUAUCCCGAUCCGGAAAGGUUCGAUCCCAGUCGCUUUGAUCCGG .......((((((((((..(((..((((((....))))))...)))....)))))))))).....((((..((......))..))))((((..((..((((....))))..))..)))). ( -36.80) >DroYak_CAF1 63443 120 - 1 CCAACACAGAUAUUGUGCUGGACAAGGGCGUCCUUGCCCUGAUUCCCGUCCAUAAUAUUCAUCACGACCCUGAGAUAUAUCCCGAACCGGAAAAGUUCGAUCCCAGUCGCUUUGAUCCGG ........(((((((((..(((..((((((....))))))...)))....)))))))))...........................(((((..((..((((....))))..))..))))) ( -30.00) >consensus CCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCUGAUUCCCGUGCACAAUAUCCACCACGAUCCGGAGAUAUAUCCCGAGCCGGAAAAGUUCGAUCCCAGUCGCUUCGAUCCGG ........((((((((((.(((..((((((....))))))...)))...))))))))))........((.(((......))).)).(((((((((..((((....))))))))..))))) (-31.84 = -33.08 + 1.24)

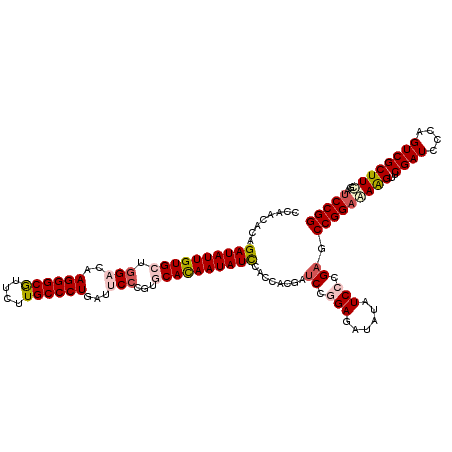
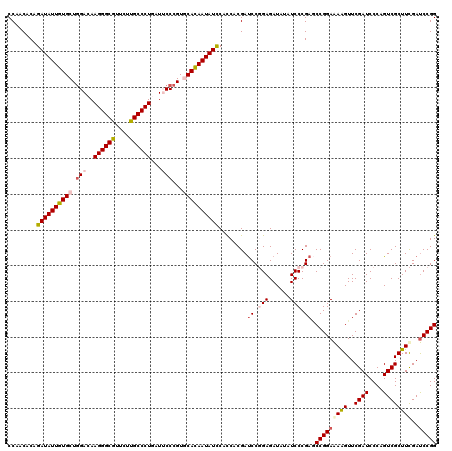
| Location | 4,076,435 – 4,076,555 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -41.14 |
| Consensus MFE | -33.06 |
| Energy contribution | -34.98 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4076435 120 + 20766785 AUAUAUCUCCGGAUCGUGGUGGAUAUUAUGCACCGGAAUCAGGGCAAGAAUGCCCUUGUCUAGCACAAUAUCUGAGUUGGGAAUUUGGUAGUCUUUAGUAGUCUCUCGAAUGAGAUGGGG ......(((((..((((...(((((((.(((...(((...((((((....))))))..))).))).)))))))...((((((..(((.(((...))).)))..))))))))))..))))) ( -35.10) >DroSec_CAF1 59198 120 + 1 AUAUAUCUCCGGAUCGUGGUGGAUAUUGUGCACGGGUAUGAGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUGGUAGUCUUUAUUAGUCUCUCGAAUGAGAUGGGG ......(((((..((((.(..((((((((((...((.(..(((((......)))))..))).))))))))))..).((((((..(((((((...)))))))..))))))))))..))))) ( -47.40) >DroSim_CAF1 67404 120 + 1 AUAUAUCUCCGGAUCGUGGUGGAUAUUGUGCACGGGUAUGAGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUGGUAGUCUUUAUUAGUCUCUCGAAUGAGAUGGGG ......(((((..((((.(..((((((((((...((.(..(((((......)))))..))).))))))))))..).((((((..(((((((...)))))))..))))))))))..))))) ( -47.40) >DroEre_CAF1 60519 120 + 1 AUAUAUCUCAGGAUCGUGGUGAAUAUUGUGAACGGGAAUCAGGGCAAGGACGCCCUUGUCCAGCACAAUAUCCGAGUGGGGAAUUUUGUAGUCUCGAUUAGCCUCCCGAAUGAGAUGGGG ...(((((((...(((.((((.((((((((....(((...(((((......)))))..)))..)))))))).).(((.((((.........)))).))).)))...))).)))))))... ( -38.60) >DroYak_CAF1 63483 120 + 1 AUAUAUCUCAGGGUCGUGAUGAAUAUUAUGGACGGGAAUCAGGGCAAGGACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUUGUAGUCUCGAUUAGCCUCUCGAACGAGAUGGGG ......((((.(((((((((....))))))..(((((..((((((......))))...(((((((((.....))))))))).....))...)))))....)))((((....)))))))). ( -37.20) >consensus AUAUAUCUCCGGAUCGUGGUGGAUAUUGUGCACGGGAAUCAGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUGGUAGUCUUUAUUAGUCUCUCGAAUGAGAUGGGG ......(((((..((((...(((((((((((...((.(..(((((......)))))..))).)))))))))))...((((((..(((((((...)))))))..))))))))))..))))) (-33.06 = -34.98 + 1.92)

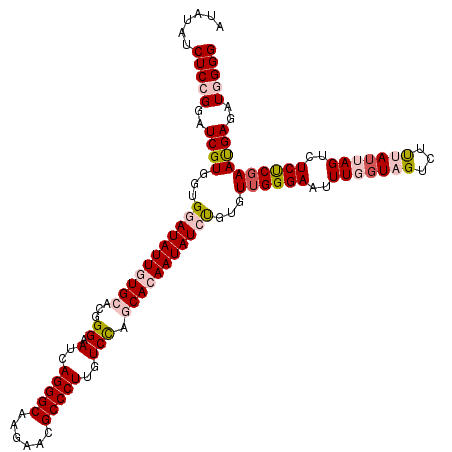
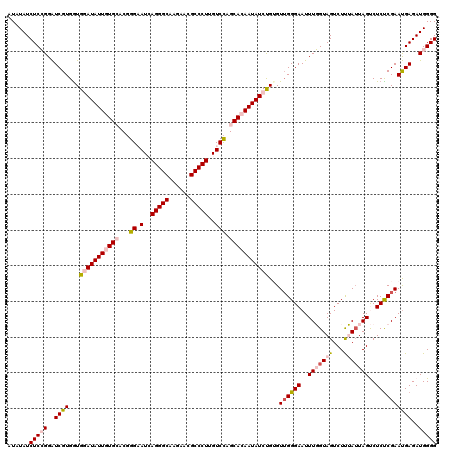
| Location | 4,076,435 – 4,076,555 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.14 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4076435 120 - 20766785 CCCCAUCUCAUUCGAGAGACUACUAAAGACUACCAAAUUCCCAACUCAGAUAUUGUGCUAGACAAGGGCAUUCUUGCCCUGAUUCCGGUGCAUAAUAUCCACCACGAUCCGGAGAUAUAU ....(((((..(((.(((..........................))).((((((((((..((..((((((....))))))...))....)))))))))).....)))....))))).... ( -31.27) >DroSec_CAF1 59198 120 - 1 CCCCAUCUCAUUCGAGAGACUAAUAAAGACUACCAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCUCAUACCCGUGCACAAUAUCCACCACGAUCCGGAGAUAUAU ....(((((..(((..((.((.....)).)).................(((((((((((((...((((((....)))))).....))).)))))))))).....)))....))))).... ( -32.90) >DroSim_CAF1 67404 120 - 1 CCCCAUCUCAUUCGAGAGACUAAUAAAGACUACCAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCUCAUACCCGUGCACAAUAUCCACCACGAUCCGGAGAUAUAU ....(((((..(((..((.((.....)).)).................(((((((((((((...((((((....)))))).....))).)))))))))).....)))....))))).... ( -32.90) >DroEre_CAF1 60519 120 - 1 CCCCAUCUCAUUCGGGAGGCUAAUCGAGACUACAAAAUUCCCCACUCGGAUAUUGUGCUGGACAAGGGCGUCCUUGCCCUGAUUCCCGUUCACAAUAUUCACCACGAUCCUGAGAUAUAU ....((((((.(((((.........(((................)))((((((((((..(((..((((((....))))))...)))....)))))))))).)).)))...)))))).... ( -37.09) >DroYak_CAF1 63483 120 - 1 CCCCAUCUCGUUCGAGAGGCUAAUCGAGACUACAAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUCCUUGCCCUGAUUCCCGUCCAUAAUAUUCAUCACGACCCUGAGAUAUAU ....(((((.(((((........)))))....................(((((((((..(((..((((((....))))))...)))....)))))))))............))))).... ( -28.60) >consensus CCCCAUCUCAUUCGAGAGACUAAUAAAGACUACCAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCUGAUUCCCGUGCACAAUAUCCACCACGAUCCGGAGAUAUAU ....(((((..(((..((.((.....)).)).................((((((((((.(((..((((((....))))))...)))...)))))))))).....)))....))))).... (-28.78 = -29.14 + 0.36)

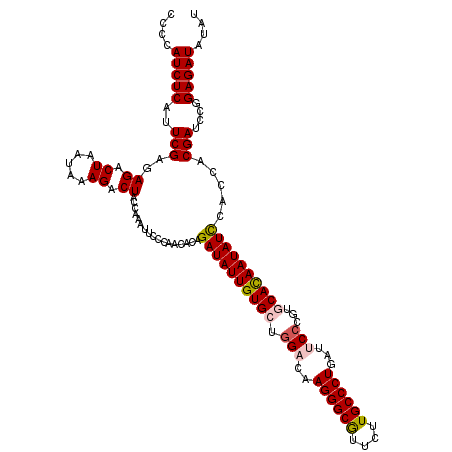
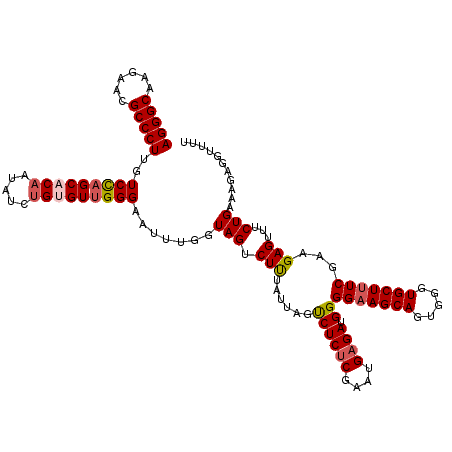
| Location | 4,076,475 – 4,076,595 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -28.54 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4076475 120 + 20766785 AGGGCAAGAAUGCCCUUGUCUAGCACAAUAUCUGAGUUGGGAAUUUGGUAGUCUUUAGUAGUCUCUCGAAUGAGAUGGGGAAGCAGUGGGUGCUUUCGAAGAGUUUCUGAAAGAGGUUUU ((((((....))))))..((((((.((.....)).))))))..((..(.(.(((((....(((((......)))))..(((((((.....)))))))))))).)..)..))......... ( -32.30) >DroSec_CAF1 59238 120 + 1 AGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUGGUAGUCUUUAUUAGUCUCUCGAAUGAGAUGGGGAAGCAGUGGGUGCUUUCGAAGAGUUUCUGAAAGUGGUUUU (((((......)))))..(((((((((.....)))))))))..((..(.(.(((((....(((((......)))))..(((((((.....)))))))))))).)..)..))......... ( -38.60) >DroSim_CAF1 67444 120 + 1 AGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUGGUAGUCUUUAUUAGUCUCUCGAAUGAGAUGGGGAAGCAGUGGGUGCUUUCGAAGAGUUUCUGAAAGUUGUUUU (((((......)))))..(((((((((.....)))))))))..((..(.(.(((((....(((((......)))))..(((((((.....)))))))))))).)..)..))......... ( -38.60) >DroEre_CAF1 60559 120 + 1 AGGGCAAGGACGCCCUUGUCCAGCACAAUAUCCGAGUGGGGAAUUUUGUAGUCUCGAUUAGCCUCCCGAAUGAGAUGGGGAAGCAGGGGAUGCUUUCGAAGAGUUUCUGAAAGAGGUUUU .((((((((....)))))))).((((((..(((......)))...)))).)).......((((((......(((((..(((((((.....))))))).....))))).....)))))).. ( -35.80) >DroYak_CAF1 63523 120 + 1 AGGGCAAGGACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUUGUAGUCUCGAUUAGCCUCUCGAACGAGAUGGGGCAGCAGGGGGUGCUUUCGAAGAGUUUCUGAAAGAUGUUUU .((((((((....)))))))).....(((((((...(..(((((((((((.((((.....(((((((......)).)))))....)))).)))......))))))))..).))))))).. ( -41.60) >consensus AGGGCAAGAACGCCCUUGUCCAGCACAAUAUCUGUGUUGGGAAUUUGGUAGUCUUUAUUAGUCUCUCGAAUGAGAUGGGGAAGCAGUGGGUGCUUUCGAAGAGUUUCUGAAAGAGGUUUU (((((......)))))..(((((((((.....))))))))).......(((.(((......((((((....)))).))(((((((.....)))))))...)))...)))........... (-28.54 = -29.10 + 0.56)

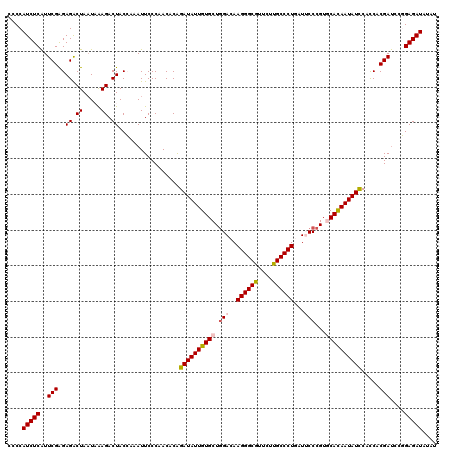
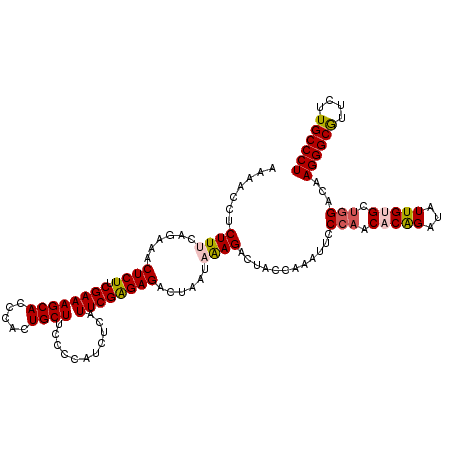
| Location | 4,076,475 – 4,076,595 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -20.54 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4076475 120 - 20766785 AAAACCUCUUUCAGAAACUCUUCGAAAGCACCCACUGCUUCCCCAUCUCAUUCGAGAGACUACUAAAGACUACCAAAUUCCCAACUCAGAUAUUGUGCUAGACAAGGGCAUUCUUGCCCU ......(((((.((...((((.((((((((.....))))...........))))))))....)))))))(((((((..((........))..))).).)))...((((((....)))))) ( -20.90) >DroSec_CAF1 59238 120 - 1 AAAACCACUUUCAGAAACUCUUCGAAAGCACCCACUGCUUCCCCAUCUCAUUCGAGAGACUAAUAAAGACUACCAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCU .......((((......((((.((((((((.....))))...........))))))))......))))............(((.(((((...))))).)))...((((((....)))))) ( -24.50) >DroSim_CAF1 67444 120 - 1 AAAACAACUUUCAGAAACUCUUCGAAAGCACCCACUGCUUCCCCAUCUCAUUCGAGAGACUAAUAAAGACUACCAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCU .......((((......((((.((((((((.....))))...........))))))))......))))............(((.(((((...))))).)))...((((((....)))))) ( -24.50) >DroEre_CAF1 60559 120 - 1 AAAACCUCUUUCAGAAACUCUUCGAAAGCAUCCCCUGCUUCCCCAUCUCAUUCGGGAGGCUAAUCGAGACUACAAAAUUCCCCACUCGGAUAUUGUGCUGGACAAGGGCGUCCUUGCCCU .........(((((...(((..(....)........(((((((.(......).))))))).....)))..(((((...(((......)))..))))))))))..((((((....)))))) ( -31.20) >DroYak_CAF1 63523 120 - 1 AAAACAUCUUUCAGAAACUCUUCGAAAGCACCCCCUGCUGCCCCAUCUCGUUCGAGAGGCUAAUCGAGACUACAAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUCCUUGCCCU ....................(((((.((((.....))))(((...((((....)))))))...)))))............(((.(((((...))))).)))...((((((....)))))) ( -29.70) >consensus AAAACCUCUUUCAGAAACUCUUCGAAAGCACCCACUGCUUCCCCAUCUCAUUCGAGAGACUAAUAAAGACUACCAAAUUCCCAACACAGAUAUUGUGCUGGACAAGGGCGUUCUUGCCCU .......((((......((((.((((((((.....))))...........))))))))......))))............(((.(((((...))))).)))...((((((....)))))) (-20.54 = -21.02 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:27 2006