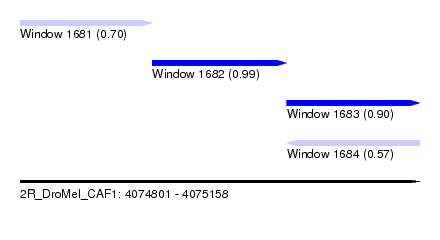
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,074,801 – 4,075,158 |
| Length | 357 |
| Max. P | 0.992176 |

| Location | 4,074,801 – 4,074,919 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.38 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4074801 118 + 20766785 UGACCUUUCGGCCACAAUAUGCGCUCGUUAGUCAAUGUUUAGCAAUUUGUUAUCAAUCAAUAAUAAAAGUAAAUAACAUAAUAAUGAGAACACAGCCGAUGGA--CCCACUCCCCCGAAG ...((..(((((.......((..(((((((....(((((......((((((((......)))))))).......)))))..)))))))..))..))))).)).--............... ( -21.82) >DroSec_CAF1 57486 118 + 1 UGACCUUUCGGCCACAAUAUGCGCUCGUUAGUCAAUGUUUAGCAAUUUGUUAUCAAUCAAUAAUAAAUGUAAAUAACAUAAUAAUGAGAACACAGGCGAUGGA--CCCACUCCCCCGAAG ......(((((........((..(((((((....(((((.....(((((((((......)))))))))......)))))..)))))))..))..((.((((..--..)).))))))))). ( -24.20) >DroSim_CAF1 65690 120 + 1 UGACCUUUCGGCCACAAUAUGCGCUCGUUAGUCAAUGUUUAGCAAUUUGUUAUCAAUCAAUAAUAAAUGUAAAUAACAUAAUAAUGAGAACACAGGCGAUGGACACCCACUCCCCCGAAG ......(((((........((..(((((((....(((((.....(((((((((......)))))))))......)))))..)))))))..))..((.(((((....))).))))))))). ( -26.10) >DroEre_CAF1 58942 118 + 1 UGACCUUUCGGCCACAAUAUGCGCUCGUUAGUCAAUGUUCAGCAAUUUGUUAUCAAUCAAUAAUAAAUGUAAAUAACAUAAUAAUGAGAGCACAGGCGCUGGG--GCCACUCCCCCGAAG ......(((((((......(((.(((((((....(((((.....(((((((((......)))))))))......)))))..))))))).)))..)))...(((--(.....)))))))). ( -30.30) >DroYak_CAF1 61867 118 + 1 UGACCUUUCGGCCACAAUAUGCGCUCGUUAGUCAAUGCUUAGCAAUUUGUUAUCAAUCAAUAAUAAAUGUAAAUAACAUAAUAAUGAGAACACAGGCGUUGGA--CCCACUCCCCCGAAG ......(((((........((..(((((((....(((.(((...(((((((((......))))))))).....))))))..)))))))..))..((.((.(..--..))).)).))))). ( -21.10) >consensus UGACCUUUCGGCCACAAUAUGCGCUCGUUAGUCAAUGUUUAGCAAUUUGUUAUCAAUCAAUAAUAAAUGUAAAUAACAUAAUAAUGAGAACACAGGCGAUGGA__CCCACUCCCCCGAAG ......(((((........((..(((((((....(((((.....(((((((((......)))))))))......)))))..)))))))..))..((.(.(((....)))).)).))))). (-22.36 = -22.96 + 0.60)

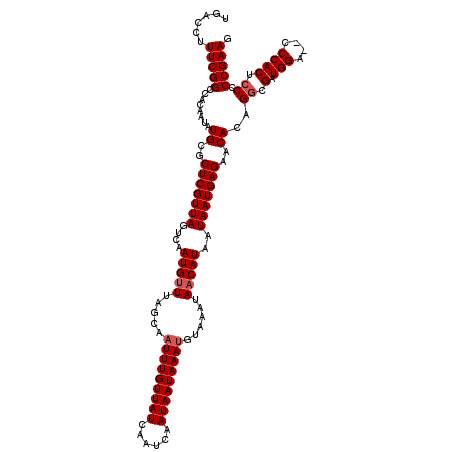
| Location | 4,074,919 – 4,075,039 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -29.50 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4074919 120 + 20766785 AUGAUGAGGCAUCAGGGCAUUGGCUCAAAUUGUCUGUGGGAAAAAUAGGAAUUUGGCCUAAAUGAGUUAUGGCCCGCAUAAACAAAAAAGGAAGUGCGAAAGCGCCCACGGAGGCAUGCG .(((((...)))))((((..(((((((.........((((..((((....))))..))))..)))))))..))))((((................((....))(((......))))))). ( -35.40) >DroSec_CAF1 57604 120 + 1 AUGAUGAGGCAGCAGGGCAUUGGCUCAAAUUGUCUGUGGGAAAAAUAGGAAUUUGGCCUAAAUGAGUUAUGGCCCGCAUAAACAAAAAAGGAAGUGCGAAAGCGCCCACGGAGGCAUGCG ........(((((.((((....))))......((((((((..............((((............))))(((((...(......)...)))))......)))))))).)).))). ( -36.00) >DroSim_CAF1 65810 112 + 1 AUGAUGAGG--------CAUUGGCUCAAAUUGUCUGUGGGAAAAAUAGGAAUUUGGCCUAAAUGAGUUAUGGCCCGCAUAAACAAAAAAGGAAGUGCGAAAGCGCCCACGGAGGCAUGCG ........(--------(((.(.(((...((((.((((.(..((((....))))((((............))))).)))).))))....((..((((....))))))...))).))))). ( -31.10) >DroEre_CAF1 59060 118 + 1 AUGAUGAGGCAUCAGGACAUUGGCUCAAAUUGUCUGUGGA-AGAAAAGGAAUUUGGCCUAAAUGAGUUAUGGCCCGCAUGAACAAAAA-GGAAGUGCGAAAGCGCCCACGGAGGCAUGCG .......(((.......(((((((.((((((.(((.....-.....))))))))))))..)))).......)))((((((........-((..((((....)))))).......)))))) ( -31.30) >DroYak_CAF1 61985 120 + 1 AUGAUGAGGCAUCAGGGCAUGGGCUCAAAUGGCCUGUGGAAAGAAAAGGAAUUUGGCCUAAAUGAGUUAUUGCCCGCAUAAACAAAAACGGAAGUGCGAAAGCGCCCACGGAGGCAUGCG .(((((...)))))(((((..((((((...((((....................))))....))))))..)))))((((........((....))((....))(((......))))))). ( -41.25) >consensus AUGAUGAGGCAUCAGGGCAUUGGCUCAAAUUGUCUGUGGGAAAAAUAGGAAUUUGGCCUAAAUGAGUUAUGGCCCGCAUAAACAAAAAAGGAAGUGCGAAAGCGCCCACGGAGGCAUGCG .......(((.......(((.(((.((((((.(((((.......))))))))))))))...))).......)))(((((................((....))(((......)))))))) (-29.50 = -30.14 + 0.64)

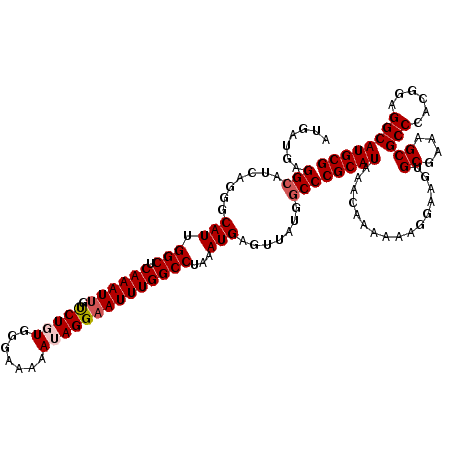
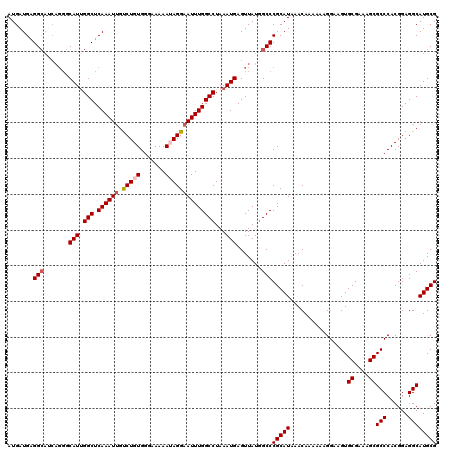
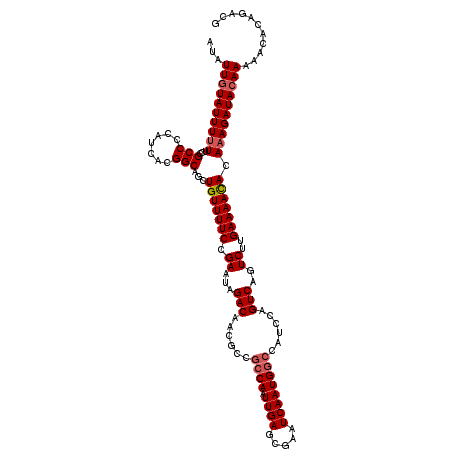
| Location | 4,075,039 – 4,075,158 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4075039 119 + 20766785 AUAUUGUAUUU-UUCCGCCCCAUCACGGCAGCUGUUUUCCGAAUGGACAACGCCACCAAUUGAGCGAAUCAAUGGCCAUCCAGUCAGUCUUGAAAACACAAAGAUACAAAAACACAGACG ...((((((((-((..(((.......))).(.(((((((.((.((((....((((....((((.....))))))))..)))).....))..))))))))))))))))))........... ( -26.80) >DroSec_CAF1 57724 120 + 1 AUAUUGUAUUUUUUCCGCCCCAUCACGGCAGCUGUUUUCCGAAUAGACAACGCCGCCAAUUGAGCGAAUCAAUGGCCAUCCAGUCAGUCUUGAAAACACAAAGAUACAAAAACACAGACU ...((((((((((...(((.......))).(.(((((((.((...(((......((((.((((.....))))))))......)))..))..))))))))))))))))))........... ( -27.80) >DroSim_CAF1 65922 120 + 1 AUAUUGUAUUUUUUCCGCCCCAUCACGGCAGCUGUUUUCCGAAUAGACAACGCCGCCAAUUGAGCGAAUCAAUGGCCAUCCAGUCAGUCUUGAAAACACAAAGAUACAAAAACACAGACG ...((((((((((...(((.......))).(.(((((((.((...(((......((((.((((.....))))))))......)))..))..))))))))))))))))))........... ( -27.80) >DroEre_CAF1 59178 120 + 1 AUAUUGUAUUUUUUCCGCCCCAUCACGGCAGCUGUUUUCCGAAUAGACAACGCCGCCAAUUGAGCGAAUCAAUGACCAGCCAGUCAGUCUUGAAAACAAAAAGAUACAAAAACACGGACG ...(((((((((((..(((.......)))...(((((((.....((((.....((((....).)))......((((......)))))))).))))))))))))))))))........... ( -27.40) >DroYak_CAF1 62105 120 + 1 AUAUUAUAUUUUUUCCGCCCCAUCACGGCAGCUGUUUUCCGAAUAGACAACUCCGCCAAUUGAGCGAAUCAAUGGCCGUCCAGUCAGUCUUGAAAAUACAAAGAUACAAAAACACGGAGG ............(((((((.......))).(.((((((.......(((....(.((((.((((.....)))))))).)....))).(((((.........)))))...))))))))))). ( -24.30) >consensus AUAUUGUAUUUUUUCCGCCCCAUCACGGCAGCUGUUUUCCGAAUAGACAACGCCGCCAAUUGAGCGAAUCAAUGGCCAUCCAGUCAGUCUUGAAAACACAAAGAUACAAAAACACAGACG ...((((((((((...(((.......)))...(((((((.((...(((......((((.((((.....))))))))......)))..))..))))))).))))))))))........... (-23.44 = -24.08 + 0.64)

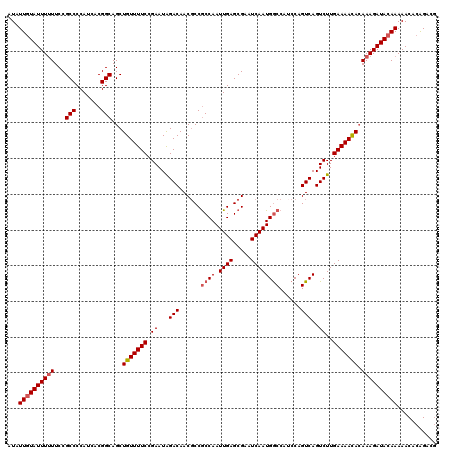
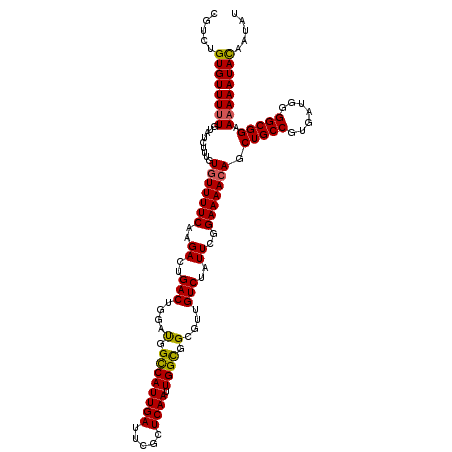
| Location | 4,075,039 – 4,075,158 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -30.08 |
| Energy contribution | -29.84 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4075039 119 - 20766785 CGUCUGUGUUUUUGUAUCUUUGUGUUUUCAAGACUGACUGGAUGGCCAUUGAUUCGCUCAAUUGGUGGCGUUGUCCAUUCGGAAAACAGCUGCCGUGAUGGGGCGGAA-AAAUACAAUAU ....(((((((((.........(((((((.....(((.(((((.(((((..(((.....)))..)))))...))))).)))))))))).(((((.......)))))))-))))))).... ( -36.80) >DroSec_CAF1 57724 120 - 1 AGUCUGUGUUUUUGUAUCUUUGUGUUUUCAAGACUGACUGGAUGGCCAUUGAUUCGCUCAAUUGGCGGCGUUGUCUAUUCGGAAAACAGCUGCCGUGAUGGGGCGGAAAAAAUACAAUAU ....(((((((((...((..((.((..((.((.....)).))..))))..))(((((((...((((((((((.(((....))).))).))))))).....)))))))))))))))).... ( -36.40) >DroSim_CAF1 65922 120 - 1 CGUCUGUGUUUUUGUAUCUUUGUGUUUUCAAGACUGACUGGAUGGCCAUUGAUUCGCUCAAUUGGCGGCGUUGUCUAUUCGGAAAACAGCUGCCGUGAUGGGGCGGAAAAAAUACAAUAU ....(((((((((...((..((.((..((.((.....)).))..))))..))(((((((...((((((((((.(((....))).))).))))))).....)))))))))))))))).... ( -36.40) >DroEre_CAF1 59178 120 - 1 CGUCCGUGUUUUUGUAUCUUUUUGUUUUCAAGACUGACUGGCUGGUCAUUGAUUCGCUCAAUUGGCGGCGUUGUCUAUUCGGAAAACAGCUGCCGUGAUGGGGCGGAAAAAAUACAAUAU .....((((((((..............((((((((........)))).))))(((((((...((((((((((.(((....))).))).))))))).....)))))))))))))))..... ( -36.70) >DroYak_CAF1 62105 120 - 1 CCUCCGUGUUUUUGUAUCUUUGUAUUUUCAAGACUGACUGGACGGCCAUUGAUUCGCUCAAUUGGCGGAGUUGUCUAUUCGGAAAACAGCUGCCGUGAUGGGGCGGAAAAAAUAUAAUAU ...................(((((((((.....((((.((((((((......((((((.....)))))))))))))).)))).......(((((.......)))))..)))))))))... ( -29.30) >consensus CGUCUGUGUUUUUGUAUCUUUGUGUUUUCAAGACUGACUGGAUGGCCAUUGAUUCGCUCAAUUGGCGGCGUUGUCUAUUCGGAAAACAGCUGCCGUGAUGGGGCGGAAAAAAUACAAUAU .....((((((((.........(((((((..((..(((....(.((((((((.....)))).)))).)....)))..))..))))))).(((((.......))))).))))))))..... (-30.08 = -29.84 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:21 2006