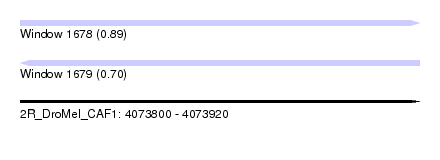
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,073,800 – 4,073,920 |
| Length | 120 |
| Max. P | 0.894548 |

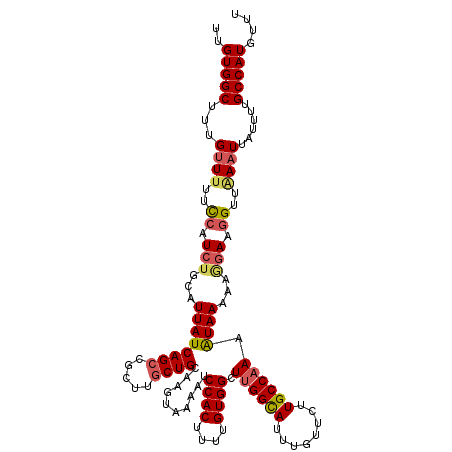
| Location | 4,073,800 – 4,073,920 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073800 120 + 20766785 AAACAUGGCAAAAUAAUUUAACCUUUCUUUUUAUUUUGGCAAGAACAAAUGCCAAACCACAAAAGUGGAUUUUACUUGCAGCAACCGGCUGAUAAUGCCGAUGGAAAAGCAAAGCCACAA .....(((((((((....................(((((((........)))))))((((....)))))))))..((((..(...((((.......))))...)....)))).))))... ( -28.60) >DroSec_CAF1 56484 120 + 1 AAACAUGGCAAAAUAAUUCAACCUUCCUUUUUACUUUGGCAAGAACAAAUGCCAAACCACAAAAGUGGAUUUUACUUGCAGCAAGCGGCUGAUAAUGCAGAUGAAAAAACAAAGCCACAA .....((((.........................(((((((........)))))))((((....)))).(((((.(((((.((......))....))))).))))).......))))... ( -25.20) >DroSim_CAF1 64703 120 + 1 AAACAUGGCAAAAUAAUUCAACCUUCCGUUUUAUUUUGGCAAGAACAAAUGCCAAGCCACAAAAGUGGAUUUUACUUGCAGCAAGCGGCUGAUAAUGCAGAUGGAAAAACAAAGCCACAA .....((((.((....)).....((((((((((((((((((........)))))).((((....))))..........((((.....))))..)))).)))))))).......))))... ( -25.40) >DroEre_CAF1 57939 119 + 1 GAACAUGGCGAAAUAAUUUAACCUUCUUUUUUAUUUGGGCAAGAACAAAUACCAAGCCACAAAAGUGGAUUUUACUUGCAGCA-GCGCCUGAUAAUGCAGAUGGGAAAACGAAGCCACAA .....((((.......(((..((.(((...(((.(..(((................((((....)))).........((....-)))))..))))...))).))..)))....))))... ( -24.00) >DroYak_CAF1 60840 120 + 1 AAACAUGGCAAAAUAAUUGAACGUUCUUUUUUAUUUUGGCAAGUACAAAUACCAAGCCACAAAAGUGGAUUUUACUUACAGCAAGCGGCUGAUAAUGCAGAUGGAAAAACAAAGCCACAA ..((.((.((((((((..((......))..)))))))).)).)).......(((..((((....))))..........((((.....))))..........)))................ ( -22.30) >consensus AAACAUGGCAAAAUAAUUCAACCUUCCUUUUUAUUUUGGCAAGAACAAAUGCCAAGCCACAAAAGUGGAUUUUACUUGCAGCAAGCGGCUGAUAAUGCAGAUGGAAAAACAAAGCCACAA .....(((((((((....................(((((((........)))))))((((....))))))))).....((((.....))))......................))))... (-19.70 = -20.10 + 0.40)

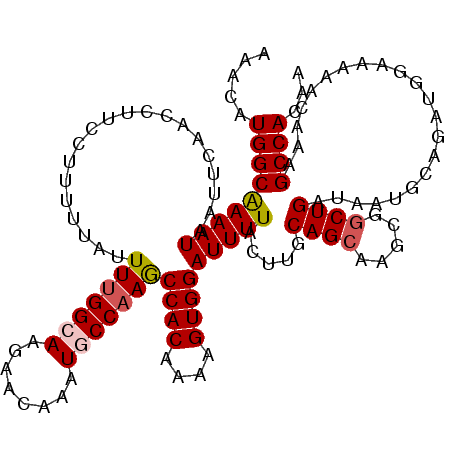
| Location | 4,073,800 – 4,073,920 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.66 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073800 120 - 20766785 UUGUGGCUUUGCUUUUCCAUCGGCAUUAUCAGCCGGUUGCUGCAAGUAAAAUCCACUUUUGUGGUUUGGCAUUUGUUCUUGCCAAAAUAAAAAGAAAGGUUAAAUUAUUUUGCCAUGUUU ..(((((...(((((((.((((((.......))))))...............((((....))))(((((((........))))))).......)))))))...........))))).... ( -31.74) >DroSec_CAF1 56484 120 - 1 UUGUGGCUUUGUUUUUUCAUCUGCAUUAUCAGCCGCUUGCUGCAAGUAAAAUCCACUUUUGUGGUUUGGCAUUUGUUCUUGCCAAAGUAAAAAGGAAGGUUGAAUUAUUUUGCCAUGUUU ..(((((..(((..........)))...((((((.(((..(((.........((((....))))(((((((........))))))))))..)))...))))))........))))).... ( -31.90) >DroSim_CAF1 64703 120 - 1 UUGUGGCUUUGUUUUUCCAUCUGCAUUAUCAGCCGCUUGCUGCAAGUAAAAUCCACUUUUGUGGCUUGGCAUUUGUUCUUGCCAAAAUAAAACGGAAGGUUGAAUUAUUUUGCCAUGUUU ..(((((...((((..((.((((..((((((((.....))))..........((((....)))).((((((........)))))).))))..)))).))..))))......))))).... ( -30.60) >DroEre_CAF1 57939 119 - 1 UUGUGGCUUCGUUUUCCCAUCUGCAUUAUCAGGCGC-UGCUGCAAGUAAAAUCCACUUUUGUGGCUUGGUAUUUGUUCUUGCCCAAAUAAAAAAGAAGGUUAAAUUAUUUCGCCAUGUUC ..(((((...((((..((.(((...((((..((.((-....(((((((.((.((((....)))).))..)))))))....))))..))))...))).))..))))......))))).... ( -30.60) >DroYak_CAF1 60840 120 - 1 UUGUGGCUUUGUUUUUCCAUCUGCAUUAUCAGCCGCUUGCUGUAAGUAAAAUCCACUUUUGUGGCUUGGUAUUUGUACUUGCCAAAAUAAAAAAGAACGUUCAAUUAUUUUGCCAUGUUU ..(((((((((((((..((..((((.((((((((((.......((((.......))))..))))).)))))..))))..))..))))))))...((....)).........))))).... ( -25.80) >consensus UUGUGGCUUUGUUUUUCCAUCUGCAUUAUCAGCCGCUUGCUGCAAGUAAAAUCCACUUUUGUGGCUUGGCAUUUGUUCUUGCCAAAAUAAAAAGGAAGGUUAAAUUAUUUUGCCAUGUUU ..(((((...((((..((.(((...((((((((.....))))..........((((....)))).((((((........)))))).))))...))).))..))))......))))).... (-22.14 = -22.66 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:16 2006