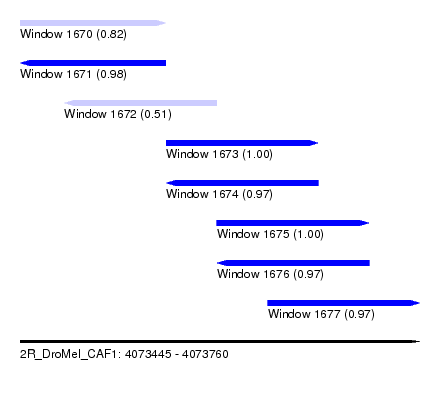
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,073,445 – 4,073,760 |
| Length | 315 |
| Max. P | 0.999754 |

| Location | 4,073,445 – 4,073,560 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -25.95 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073445 115 + 20766785 AUUUGUUUUGGGGACUUGCCA-----AAAAAUUGUAAAGCAGACAAGCGCCCUUUGACAUCUCGUUUCGCGUUGGCUUUUUAUCAAAUCAGGACUUGAUUUGAUUAGACCCCGAACGGAA .(((((((.((((....((((-----.....((((.......))))((((....(((....)))....)))))))).(((.((((((((((...)))))))))).)))))))))))))). ( -31.10) >DroSec_CAF1 56139 115 + 1 AUUUGUUCUGGGCACUUGCCA-----AAAAAUUGUAAAGCAGACAAGCGCCCUUUGACAUCUCGUUUCGCGUUGGCUUUUUAUCAAAUCGGGACUUGAUUUGAUUAGACCCCGAACGGAA .(((((((.((((....((((-----.....((((.......))))((((....(((....)))....)))))))).....(((((((((.....)))))))))..).))).))))))). ( -29.60) >DroSim_CAF1 64358 115 + 1 AUUUGUUCUGGGGACUUGCCA-----AAAAAUUGUAAAGCAGACAAGCGCCCUUUGACAUCUCGUUUCGCGUUGGCUUUUUAUCAAAUCAGGACUUGAUUUGAUUAGACCCCGAACGGAA .(((((((.((((....((((-----.....((((.......))))((((....(((....)))....)))))))).(((.((((((((((...)))))))))).)))))))))))))). ( -33.20) >DroEre_CAF1 57624 93 + 1 AUUUGUCCUGGGGACUUGCCG-----AAAAAUUGUAAAGCAGACUAG----------------------ACUUGGCUUCUUAUCAAAUCAGGACUUGAUUUGAUUAGACCCCGAACGGAA .(((((..(((((....((((-----(....(((.....))).....----------------------..))))).(((.((((((((((...)))))))))).)))))))).))))). ( -25.80) >DroYak_CAF1 60497 120 + 1 AUUUGUUCUGGCGACUUGCCAGAAACAAAAAUUGUUAAGCAGACAAGCGCCCUUUGACAUCCCGUUUCGCGUUGGCUUUUUAUCAAAUCAGGACUUGAUUUGAUUAGACCCCGAACGGAA .(((((((((((.....))))).))))))...((((((((........))...))))))..(((((.((.(..((((....((((((((((...)))))))))).)).)))))))))).. ( -33.00) >consensus AUUUGUUCUGGGGACUUGCCA_____AAAAAUUGUAAAGCAGACAAGCGCCCUUUGACAUCUCGUUUCGCGUUGGCUUUUUAUCAAAUCAGGACUUGAUUUGAUUAGACCCCGAACGGAA .(((((((.((((....(((...........(((.....)))...(((((....((......))....)))))))).(((.((((((((((...)))))))))).)))))))))))))). (-25.95 = -26.62 + 0.67)

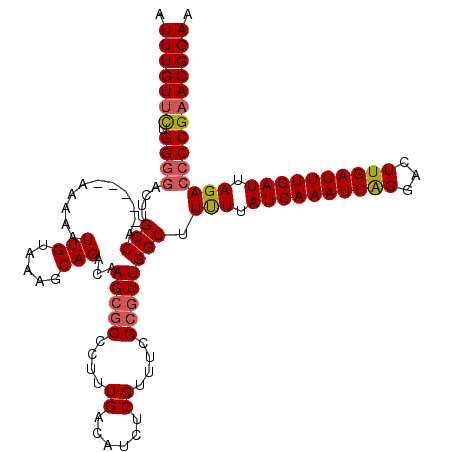
| Location | 4,073,445 – 4,073,560 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -24.52 |
| Energy contribution | -26.24 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073445 115 - 20766785 UUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCUGCUUUACAAUUUUU-----UGGCAAGUCCCCAAAACAAAU ........((((.((.(((((((((.....))))))))).....(((((.(((..(((((.((((....))))))))).)))...........)-----)))).)).))))......... ( -32.10) >DroSec_CAF1 56139 115 - 1 UUCCGUUCGGGGUCUAAUCAAAUCAAGUCCCGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCUGCUUUACAAUUUUU-----UGGCAAGUGCCCAGAACAAAU ....((((((.((((.((((((((.......)))))))).....((....))......)))).))...(((((((((((...............-----.))))))))))).)))).... ( -38.09) >DroSim_CAF1 64358 115 - 1 UUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCUGCUUUACAAUUUUU-----UGGCAAGUCCCCAGAACAAAU ....((((((((.((.(((((((((.....))))))))).....(((((.(((..(((((.((((....))))))))).)))...........)-----)))).)).)))).)))).... ( -36.20) >DroEre_CAF1 57624 93 - 1 UUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAGAAGCCAAGU----------------------CUAGUCUGCUUUACAAUUUUU-----CGGCAAGUCCCCAGGACAAAU ....((((((((.((.(((((((((.....)))))))))..(((((..((.----------------------.....))))))).........-----.....)).)))).)))).... ( -25.70) >DroYak_CAF1 60497 120 - 1 UUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGGGAUGUCAAAGGGCGCUUGUCUGCUUAACAAUUUUUGUUUCUGGCAAGUCGCCAGAACAAAU (((((((((.(((...(((((((((.....))))))))).....)))....)).)))))))(((.....((((((((((.....((((.....))))...)))))).))))...)))... ( -36.60) >consensus UUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCUGCUUUACAAUUUUU_____UGGCAAGUCCCCAGAACAAAU ....((((((((.((.(((((((((.....))))))))).....(((........(((((.((((....))))))))).((.....))............))).)).)))).)))).... (-24.52 = -26.24 + 1.72)

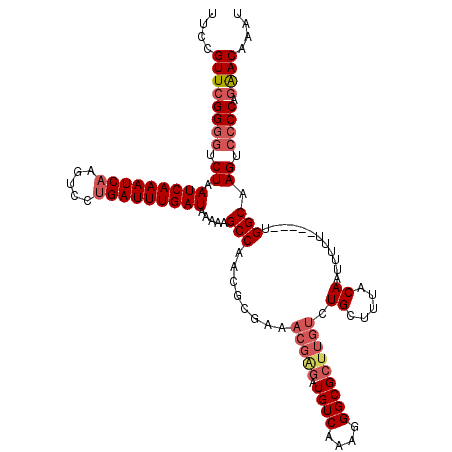
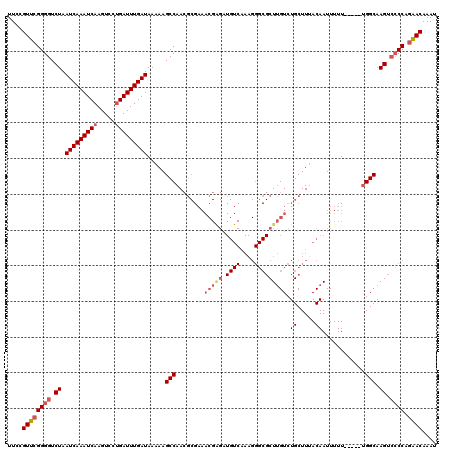
| Location | 4,073,480 – 4,073,600 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073480 120 - 20766785 CAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCAUUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCU .....((.(((((.....)))))..(((...........(((((....)))))...(((((((((.....))))))))).....)))...))...(((((.((((....))))))))).. ( -31.50) >DroSec_CAF1 56174 120 - 1 CAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCAUUCCGUUCGGGGUCUAAUCAAAUCAAGUCCCGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCU .....((.(((((.....)))))..(((.....(((((((......((((((.((..........))))))))..)))))))..)))...))...(((((.((((....))))))))).. ( -30.50) >DroSim_CAF1 64393 120 - 1 CAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCAUUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCU .....((.(((((.....)))))..(((...........(((((....)))))...(((((((((.....))))))))).....)))...))...(((((.((((....))))))))).. ( -31.50) >DroEre_CAF1 57659 98 - 1 CAUCAGCAGCUGGCCCAUUCAGCCAGCCAAUAAUUUAUCAUUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAGAAGCCAAGU----------------------CUAGUCU ........((((((.......)))))).....................((..(((.(((((((((.....))))))))).)))..))....----------------------....... ( -25.90) >DroYak_CAF1 60537 120 - 1 CACCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCAUUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGGGAUGUCAAAGGGCGCUUGUCU .((.(((..(((((.......)))))((.....(((..(((((((((((.(((...(((((((((.....))))))))).....)))....)).)))))))))..)))..))))).)).. ( -37.60) >consensus CAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCAUUCCGUUCGGGGUCUAAUCAAAUCAAGUCCUGAUUUGAUAAAAAGCCAACGCGAAACGAGAUGUCAAAGGGCGCUUGUCU .....((.((((((.......))))).).....(((((((......((((((.((..........))))))))..)))))))..)).........(((((.((((....))))))))).. (-22.62 = -23.38 + 0.76)

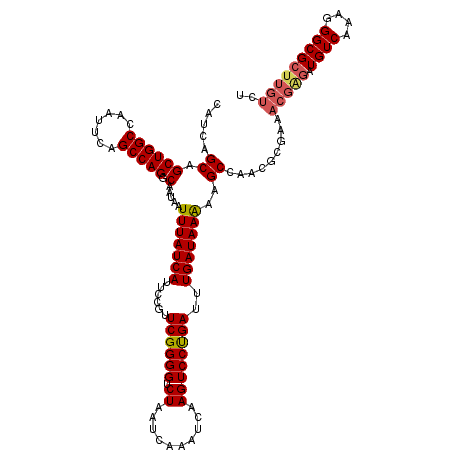
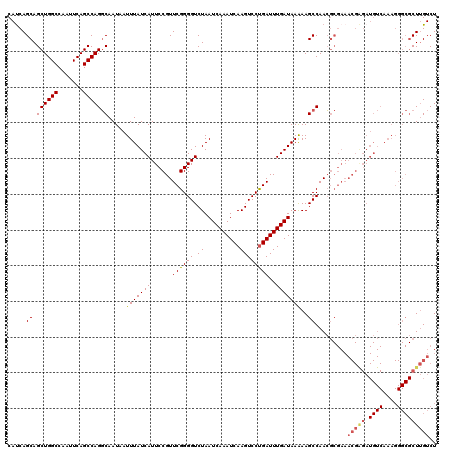
| Location | 4,073,560 – 4,073,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -34.30 |
| Energy contribution | -33.98 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073560 120 + 20766785 UGAUAAAUUAUUGCCUGGCUGAAUUGGCCAGCUGCUGAUGUCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAG .........((((..((((...((..((.....))..)).......((((((.........)))))).))))..)))).(((...(((((((((((........))))))))))).))). ( -35.20) >DroSec_CAF1 56254 120 + 1 UGAUAAAUUAUUGCCUGGCUGAAUUGGCCAGCUGCUGAUGUCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUCGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAG .........((((..((((...((..((.....))..)).......((((((.........)))))).))))..))))..((...(((((((((((........))))))))))).)).. ( -34.60) >DroSim_CAF1 64473 120 + 1 UGAUAAAUUAUUGCCUGGCUGAAUUGGCCAGCUGCUGAUGUCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUCGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAG .........((((..((((...((..((.....))..)).......((((((.........)))))).))))..))))..((...(((((((((((........))))))))))).)).. ( -34.60) >DroEre_CAF1 57717 119 + 1 UGAUAAAUUAUUGGCUGGCUGAAUGGGCCAGCUGCUGAUGUCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAGCGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCA- ....(((((...((((((((.....))))))))(((...((.....((((((.........)))))).))..))))))))((...(((((((((((........))))))))))).)).- ( -40.00) >DroYak_CAF1 60617 119 + 1 UGAUAAAUUAUUGCCUGGCUGAAUUGGCCAGCUGCUGGUGUCCUCUGACGACUUAAUGCGUGUCGUCUGCCAAACGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCA- ....(((((...((((((((.....))))))..))(((((......((((((.........)))))))))))...)))))((...(((((((((((........))))))))))).)).- ( -36.20) >consensus UGAUAAAUUAUUGCCUGGCUGAAUUGGCCAGCUGCUGAUGUCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAG .........((((..((((...((..((.....))..)).......((((((.........)))))).))))..))))..((...(((((((((((........))))))))))).)).. (-34.30 = -33.98 + -0.32)

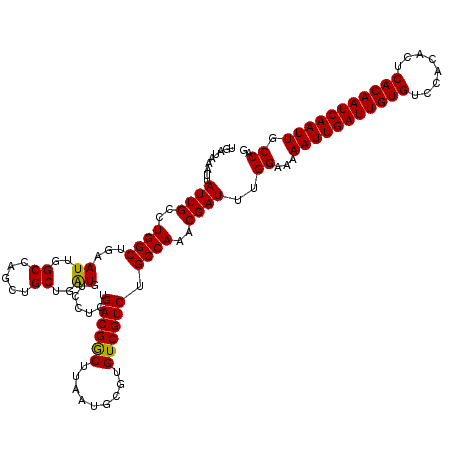
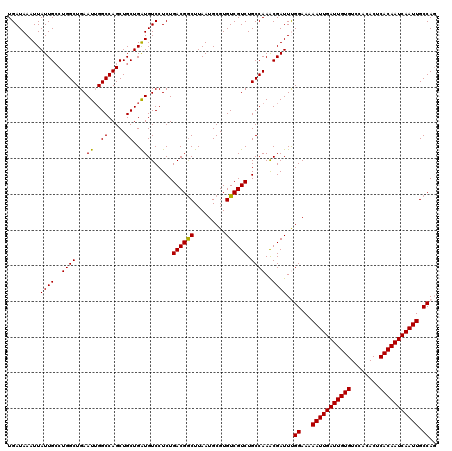
| Location | 4,073,560 – 4,073,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -31.96 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073560 120 - 20766785 CUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGACAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCA ((((((((((((((((........)))))))))))............(((((.((((....((.....)))))).........(((...))))))))....))))).............. ( -32.30) >DroSec_CAF1 56254 120 - 1 CUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCGAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGACAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCA ((((((((((((((((........))))))))))).....((((.(.(..((.((((....((.....)))))).((.....))....))..).).)))).))))).............. ( -33.50) >DroSim_CAF1 64473 120 - 1 CUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCGAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGACAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCA ((((((((((((((((........))))))))))).....((((.(.(..((.((((....((.....)))))).((.....))....))..).).)))).))))).............. ( -33.50) >DroEre_CAF1 57717 119 - 1 -UGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGCUUGGCAGACGACACGCAUUAAGCCGUCAGAGGACAUCAGCAGCUGGCCCAUUCAGCCAGCCAAUAAUUUAUCA -.((((((((((((((........)))))))))))..........((.((........)).)).....)))(((....))).......((((((.......))))))............. ( -32.90) >DroYak_CAF1 60617 119 - 1 -UGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGUUUGGCAGACGACACGCAUUAAGUCGUCAGAGGACACCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCA -(((.(((((((((((........)))))))))))...)))....(((((((.((((((.........)))))).(((....((((...))))....))).)))))))............ ( -38.70) >consensus CUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGACAUCAGCAGCUGGCCAAUUCAGCCAGGCAAUAAUUUAUCA .(((.(((((((((((........)))))))))))...)))....(((((((.((((....((.....)))))).(((....((((...))))....))).)))))))............ (-31.96 = -31.60 + -0.36)

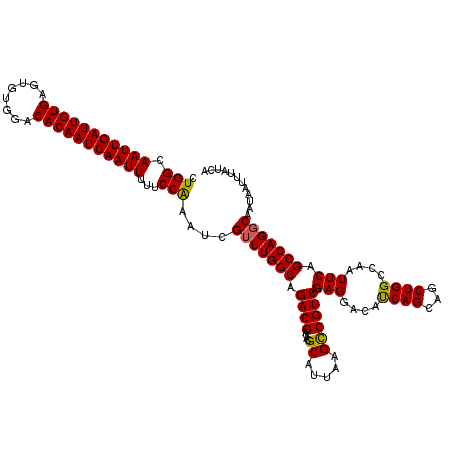
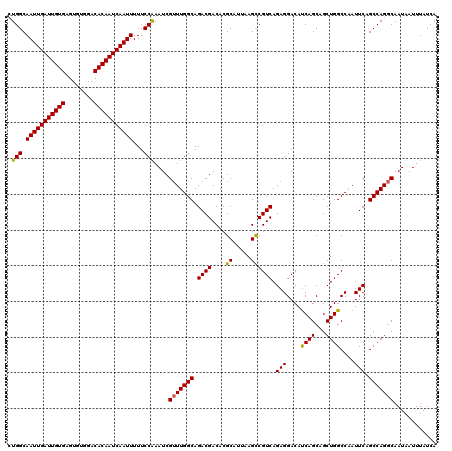
| Location | 4,073,600 – 4,073,720 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -34.90 |
| Energy contribution | -35.94 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.87 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073600 120 + 20766785 UCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGCCGAAUGGUCAAGCAAUAAACAAUU .....((((.((.....))...(((.(((((......(((((...(((((((((((........))))))))))).)))))((....)).))))).)))...)))).............. ( -39.10) >DroSec_CAF1 56294 120 + 1 UCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUCGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGUCGAAUGGCCAAGCAAUAAACAAUU ..........((((...((...(((.(((((.........((...(((((((((((........))))))))))).)).((.(....)))))))).)))...)).))))........... ( -38.50) >DroSim_CAF1 64513 120 + 1 UCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUCGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGUCGAAUGGCCAAGCAAUAAACAAUU ..........((((...((...(((.(((((.........((...(((((((((((........))))))))))).)).((.(....)))))))).)))...)).))))........... ( -38.50) >DroEre_CAF1 57757 119 + 1 UCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAGCGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCA-AAGGAAACUCGGCAGCCGAAUGGCCAAGCAAUAAACAAUU ......((((((.........))))))(((...((.((((((...(((((((((((........)))))))))))(((.-.((....)).)))..)))))).))...))).......... ( -40.20) >DroYak_CAF1 60657 119 + 1 UCCUCUGACGACUUAAUGCGUGUCGUCUGCCAAACGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCA-AAGGAAACUCGGCAGCAGAACGGCCAAGCAAUAAACAAUU ............(((.(((..(((((((((....((((((((...(((((((((((........))))))))))).)))-))(....))))...)))).)))))...))).)))...... ( -38.30) >consensus UCCUCUGACGGCUUAAUGCGUGUCGUCUGCCAAACGAUUUGGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGCCGAAUGGCCAAGCAAUAAACAAUU ..........((((...((...(((.(((((......(((((...(((((((((((........))))))))))).)))))((....)).))))).)))...)).))))........... (-34.90 = -35.94 + 1.04)

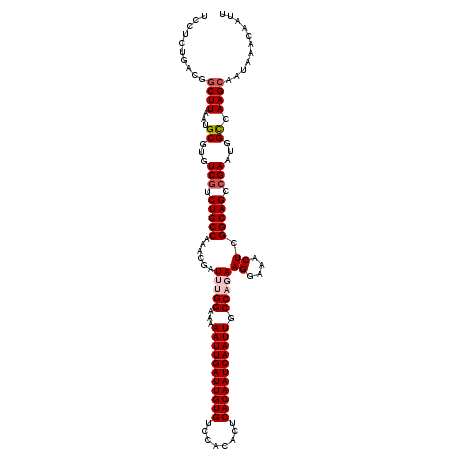
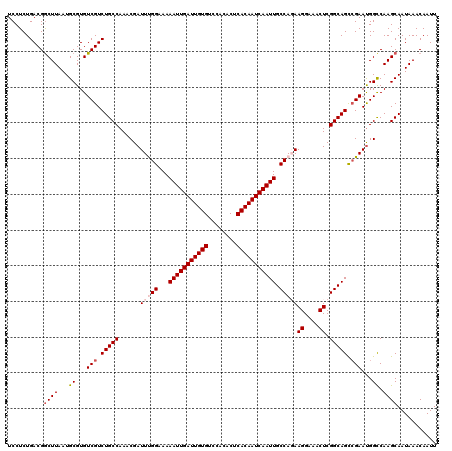
| Location | 4,073,600 – 4,073,720 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -32.18 |
| Energy contribution | -31.82 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073600 120 - 20766785 AAUUGUUUAUUGCUUGACCAUUCGGCUGCCGAGUUUCCUUCUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGA ....(((((.(((.((.....(((.((((((((........(((.(((((((((((........)))))))))))...))).....)))))))).))))).))).))))).......... ( -33.12) >DroSec_CAF1 56294 120 - 1 AAUUGUUUAUUGCUUGGCCAUUCGACUGCCGAGUUUCCUUCUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCGAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGA ...........(((((((.........)))))))...(((((((((((((((((((........)))))))))))....((..(((((.....)))))..)).........)))))))). ( -36.00) >DroSim_CAF1 64513 120 - 1 AAUUGUUUAUUGCUUGGCCAUUCGACUGCCGAGUUUCCUUCUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCGAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGA ...........(((((((.........)))))))...(((((((((((((((((((........)))))))))))....((..(((((.....)))))..)).........)))))))). ( -36.00) >DroEre_CAF1 57757 119 - 1 AAUUGUUUAUUGCUUGGCCAUUCGGCUGCCGAGUUUCCUU-UGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGCUUGGCAGACGACACGCAUUAAGCCGUCAGAGGA ....(((((.(((.((.....(((.(((((((((....((-(((.(((((((((((........)))))))))))...)))))..))))))))).))))).))).))))).......... ( -40.20) >DroYak_CAF1 60657 119 - 1 AAUUGUUUAUUGCUUGGCCGUUCUGCUGCCGAGUUUCCUU-UGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGUUUGGCAGACGACACGCAUUAAGUCGUCAGAGGA ...........(((((((.(.....).))))))).(((((-(((((((((((((((........)))))))))))........(((((.....))))).............))))))))) ( -37.70) >consensus AAUUGUUUAUUGCUUGGCCAUUCGGCUGCCGAGUUUCCUUCUGGCAAUUGAUUGUGAGUGUGGACACAAUCAAUUUUUCCAAAUCGUUUGGCAGACGACACGCAUUAAGCCGUCAGAGGA ....(((((.(((.((.....(((.((((((((........(((.(((((((((((........)))))))))))...))).....)))))))).))))).))).))))).......... (-32.18 = -31.82 + -0.36)

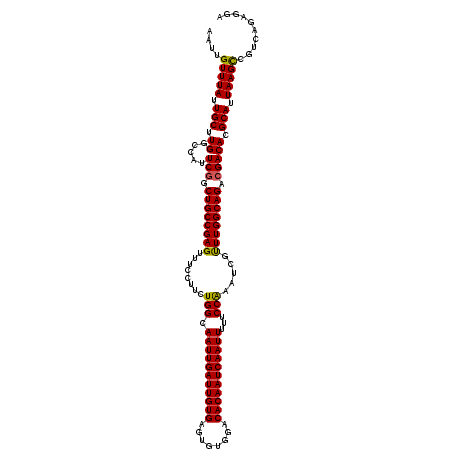
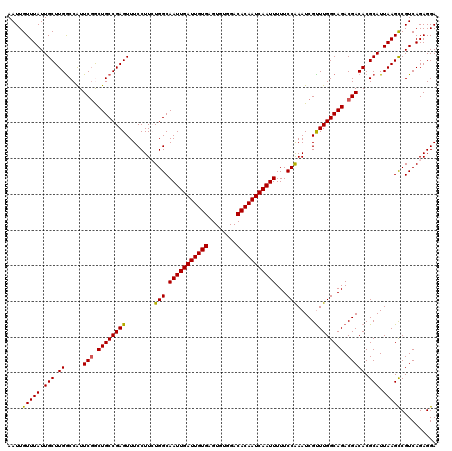
| Location | 4,073,640 – 4,073,760 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -27.64 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4073640 120 + 20766785 GGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGCCGAAUGGUCAAGCAAUAAACAAUUCCACAGCUAAAAGUGUCUGGUUUGGGCUUGGGUUCGGGAU .....(((((((((((........)))))))))))(((.((.(....))))))..((((((..((((((..(((((.....(((........)))....))))).))))))))))))... ( -38.70) >DroSec_CAF1 56334 120 + 1 GGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGUCGAAUGGCCAAGCAAUAAACAAUUCCACAGCUAAAAGUGUUUGGUUUGGGUUUGGUUUCGGGAU .....(((((((((((........)))))))))))(((.((.(....))))))..(((((..(((((((..(((((.....(((........)))....))))).))))))))))))... ( -36.40) >DroSim_CAF1 64553 120 + 1 GGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGUCGAAUGGCCAAGCAAUAAACAAUUCCACAGCUAAAAGUGUCUGGUUUGGGUUUGGGUUCGGGAU .....(((((((((((........)))))))))))(((.((.(....))))))..((((((..((((((..(((((.....(((........)))....))))).))))))))))))... ( -35.80) >DroEre_CAF1 57797 114 + 1 GGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCA-AAGGAAACUCGGCAGCCGAAUGGCCAAGCAAUAAACAAUUCCACAGCUAAAAGUGUCUGGGUUGAGUGUGGAUGC----- .............(((((((((((((........((((.-.((....)).))))(((....)))................((.((((.......).))))).)))))))))))))----- ( -34.40) >DroYak_CAF1 60697 115 + 1 GGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCA-AAGGAAACUCGGCAGCAGAACGGCCAAGCAAUAAACAAUUCCACAUCUAAAAGGGUCUGGGUUGAGUUUGGGGACG---- ((...(((((((((((........))))))))))).)).-....(((((((((.((......))................(((.((((....)))).))))))))))))(....).---- ( -33.00) >consensus GGAAAAAUUGAUUGUGUCCACACUCACAAUCAAUUGCCAGAAGGAAACUCGGCAGCCGAAUGGCCAAGCAAUAAACAAUUCCACAGCUAAAAGUGUCUGGUUUGGGUUUGGGUUCGGGAU .....(((((((((((........)))))))))))(((...((....)).)))..((((((..((((((......(((..(((..((.....))...))).))).))))))))))))... (-27.64 = -28.48 + 0.84)

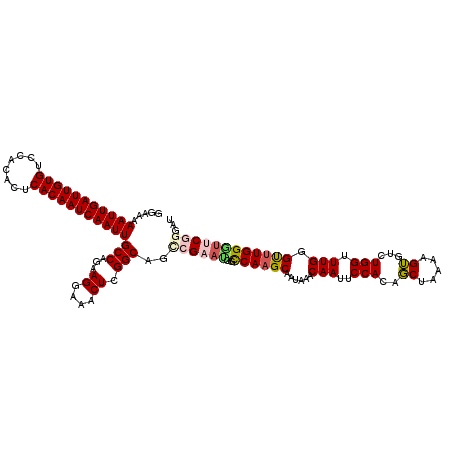
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:14 2006