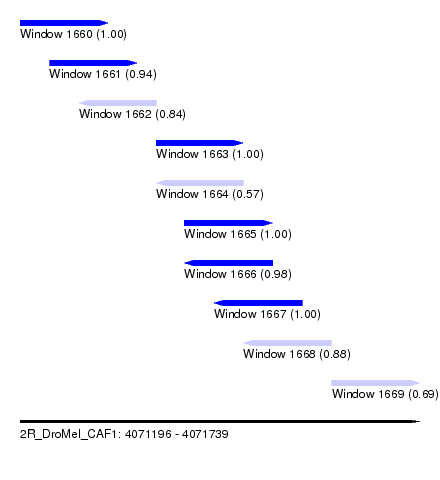
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,071,196 – 4,071,739 |
| Length | 543 |
| Max. P | 0.999611 |

| Location | 4,071,196 – 4,071,316 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -28.84 |
| Energy contribution | -29.64 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071196 120 + 20766785 AAAGUUUCCCCACACAAAAUAUAAUGUGUACGACCCUUGUCAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAAC .(((..((...(((((........)))))..))..)))......((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))......... ( -29.50) >DroSec_CAF1 53877 120 + 1 AAAGUUGCCCCACACAAAGUAUAAUGUGUACGACCCUUGUCAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAAC ...(((((...(((((........)))))..(((....)))...((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))....))))) ( -32.30) >DroSim_CAF1 62056 120 + 1 AAAGUUGCCCCACACAAAGUAUAAUGUGUACGACCCUUGUCAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAAC ...(((((...(((((........)))))..(((....)))...((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))....))))) ( -32.30) >DroEre_CAF1 55218 119 + 1 AAAGUUGCCCC-CACAAAAUAUAAUGUGUACGACCCUUGUCAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAAC ...(((((...-((((........))))...(((....)))...((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))....))))) ( -30.50) >DroYak_CAF1 57850 120 + 1 AAAGUUGCCCCACACAAAAUAUAAUGUGCACGACCCUUGUCAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGAGGCAAAUGAGCUUGGCAGGCACCAGAAAAUAAAAAGUAAC ...(.((((....((((....(((.(((((.(((....))).))))).)))...))))(((((((((.(((((((.....)))))))))).)))))))))).)................. ( -31.20) >consensus AAAGUUGCCCCACACAAAAUAUAAUGUGUACGACCCUUGUCAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAAC ...(((((...(((((........)))))..(((....)))...((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))....))))) (-28.84 = -29.64 + 0.80)

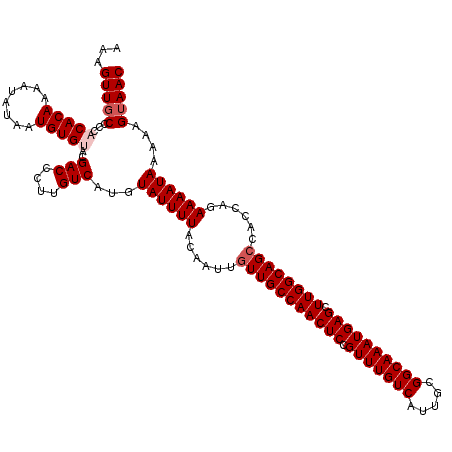
| Location | 4,071,236 – 4,071,355 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071236 119 + 20766785 CAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAACACGAGCCAAAACAAGAAACUCGCAUAAAUUACCCG-GCCA ....((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))...........(.(((...........................)-))). ( -26.03) >DroSec_CAF1 53917 120 + 1 CAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAACACGAGCCAAAACAAGAAACUCGCAUAAAUUACCCGGGCCA ....((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))..........((((.............)))).................. ( -26.02) >DroSim_CAF1 62096 120 + 1 CAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAACACGAGCCAAAACAAGAAACUCGCAUAAAUUACCCGGGCCA ....((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))..........((((.............)))).................. ( -26.02) >DroEre_CAF1 55257 120 + 1 CAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAACACGAGCCACAACAAGAAACUCGCGUAAAUUACCCGGACCA ...(((.(((((....(((((((((((.(((((((.....)))))))))).))))))))......................((((.............)))).))))).)))........ ( -29.32) >DroYak_CAF1 57890 120 + 1 CAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGAGGCAAAUGAGCUUGGCAGGCACCAGAAAAUAAAAAGUAACACGAGCCAAAACAAGAAACUCGCAUAAAUUACCCGGGCAA ....((((((.....((((((((((((.(((((((.....)))))))))).)))))).)))....)))))).............(((............................))).. ( -23.69) >consensus CAUGUAUUUUACAAUUGUUGCCAACUCCGUUUGUCAUUGCGGCAAAUGAGCUUGGCAGCCACCAGAAAAUAAAAAGUAACACGAGCCAAAACAAGAAACUCGCAUAAAUUACCCGGGCCA ....((((((......(((((((((((.(((((((.....)))))))))).))))))))......))))))..........((((.............)))).................. (-25.14 = -25.34 + 0.20)

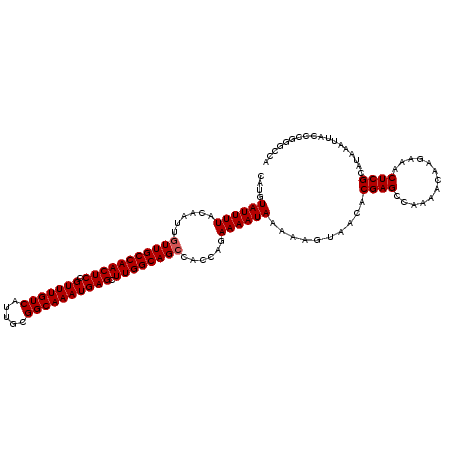
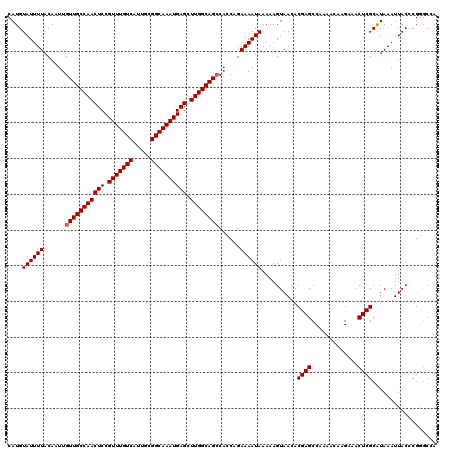
| Location | 4,071,276 – 4,071,381 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -27.11 |
| Energy contribution | -27.83 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071276 105 - 20766785 ---CUUCCCAAUUCGG-------AGAGGCAGCUGCU----UGGC-CGGGUAAUUUAUGCGAGUUUCUUGUUUUGGCUCGUGUUACUUUUUAUUUUCUGGUGGCUGCCAAGCUCAUUUGCC ---.((((......))-------)).(((((..(((----((((-(((((.......(((((...)))))....))))).((((((...........)))))).)))))))....))))) ( -31.20) >DroSec_CAF1 53957 106 - 1 ---CUUCACAAUCCGG-------CGAGGCAGCUGCG----UGGCCCGGGUAAUUUAUGCGAGUUUCUUGUUUUGGCUCGUGUUACUUUUUAUUUUCUGGUGGCUGCCAAGCUCAUUUGCC ---.....(((...((-------(..((((((..((----((((.(((((.......(((((...)))))....))))).))))).............)..))))))..)))...))).. ( -32.01) >DroSim_CAF1 62136 106 - 1 ---CUUCACAAUUCGG-------GGAGGCAGCUGCU----UGGCCCGGGUAAUUUAUGCGAGUUUCUUGUUUUGGCUCGUGUUACUUUUUAUUUUCUGGUGGCUGCCAAGCUCAUUUGCC ---.....(((....(-------((.((((((..((----.((...(((((((....(((((((.........)))))))))))))).......)).))..))))))...)))..))).. ( -29.90) >DroEre_CAF1 55297 108 - 1 AACGUUCAGAAUUUGG-------AGAGGCAGCUG-----CUGGUCCGGGUAAUUUACGCGAGUUUCUUGUUGUGGCUCGUGUUACUUUUUAUUUUCUGGUGGCUGCCAAGCUCAUUUGCC ...............(-------((.((((((..-----(..(...((((((...((((((((..(.....)..))))))))......)))))).)..)..))))))...)))....... ( -37.30) >DroYak_CAF1 57930 117 - 1 ---CUUCACAAUAUGGCAGGCAGGGAGGCAGCUGCCUAGCUUGCCCGGGUAAUUUAUGCGAGUUUCUUGUUUUGGCUCGUGUUACUUUUUAUUUUCUGGUGCCUGCCAAGCUCAUUUGCC ---..........((((((((((((((((((((....))).)))).(((((((....(((((((.........))))))))))))))......))))..))))))))).((......)). ( -39.10) >consensus ___CUUCACAAUUCGG_______AGAGGCAGCUGCU____UGGCCCGGGUAAUUUAUGCGAGUUUCUUGUUUUGGCUCGUGUUACUUUUUAUUUUCUGGUGGCUGCCAAGCUCAUUUGCC ..........................((((((((((..........(((((((....(((((((.........))))))))))))))..........))))))))))..((......)). (-27.11 = -27.83 + 0.72)

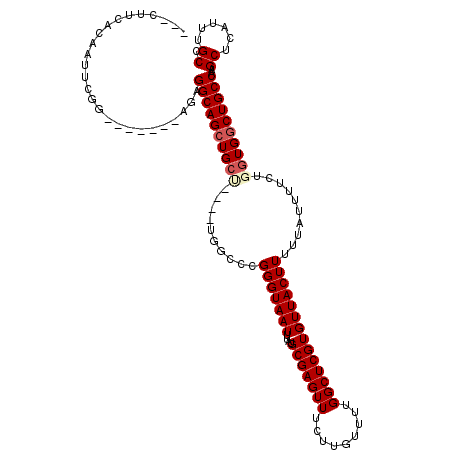
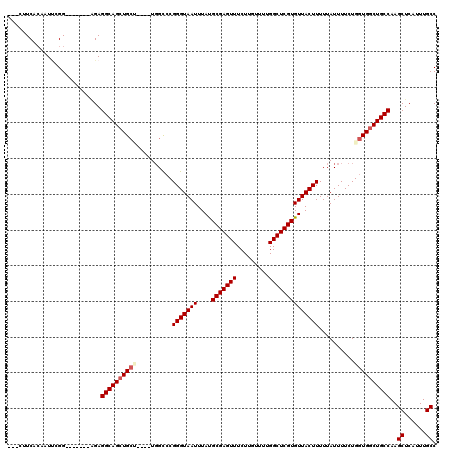
| Location | 4,071,381 – 4,071,499 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
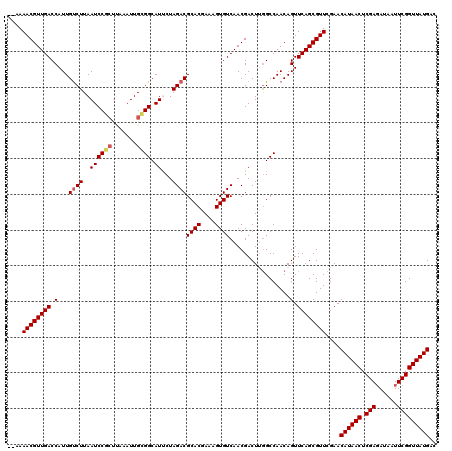
>2R_DroMel_CAF1 4071381 118 + 20766785 --AAAACGUUGACCAUUGUCUUAAUCCGCUCAAAUUGCGGCAUUCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGAC --..((((((((.(...((((.(((((((.......)))).)))..))))((((....)))).................).))))))))....((((((.((((....)))))))))).. ( -35.20) >DroSec_CAF1 54063 118 + 1 --AAAACGUUGACCAUUGUCUUAAUCCGCUUAAAUUGCGGCAUCCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGAC --..((((((((.(...((((..((((((.......)))).))...))))((((....)))).................).))))))))....((((((.((((....)))))))))).. ( -34.10) >DroSim_CAF1 62242 118 + 1 --AAAACGUUGACCAUUGUCUUAAUCCGCUUAAAUUGCGGCAUCCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGAC --..((((((((.(...((((..((((((.......)))).))...))))((((....)))).................).))))))))....((((((.((((....)))))))))).. ( -34.10) >DroEre_CAF1 55405 120 + 1 CUGAAACGUUGACCAUUGUCUUAAUCCGGCCAAAUUGUGGCAUUCUAGACGCACGAAUGUGUCAACGACUCGGCCAACAGUUCAGCGUUCGAACAUAACGCGAUAUAAUUCGGUUAUGAC (((((..((((.((...(((........((((.....))))......((((((....))))))...)))..)).))))..)))))........((((((.(((......))))))))).. ( -36.20) >DroYak_CAF1 58047 118 + 1 --CAAACGUUGACCAUUGCCUUAAUCCCCUUCAAUUGUGGCAUUCUAGACGCACGAAAGUGUCAACGACUCAGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGAC --(.((((((((.(..((((.((((........)))).)))).....(((((......)))))................).)))))))).)..((((((.((((....)))))))))).. ( -29.70) >consensus __AAAACGUUGACCAUUGUCUUAAUCCGCUUAAAUUGCGGCAUUCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGAC ....((((((((.(...((((..((((((.......)))).))...))))((((....)))).................).))))))))....((((((.(((......))))))))).. (-29.30 = -29.78 + 0.48)

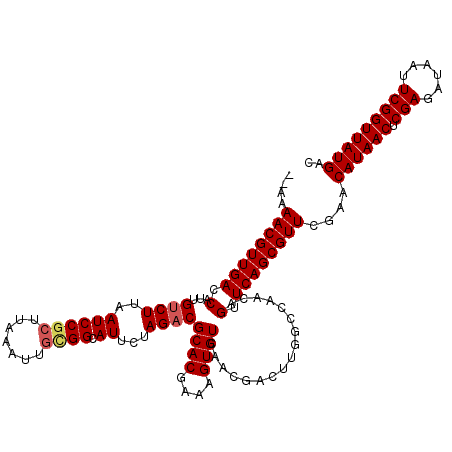
| Location | 4,071,381 – 4,071,499 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -30.26 |
| Energy contribution | -30.06 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
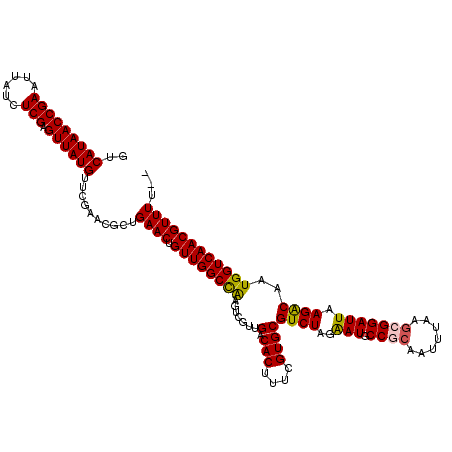
>2R_DroMel_CAF1 4071381 118 - 20766785 GUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGAAUGCCGCAAUUUGAGCGGAUUAAGACAAUGGUCAACGUUUU-- ..(((((((((......))).))))))..........((((.((((((((.......(.(((....))))((((..(((.((((.......))))))).))))..)))))))))))).-- ( -33.40) >DroSec_CAF1 54063 118 - 1 GUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGGAUGCCGCAAUUUAAGCGGAUUAAGACAAUGGUCAACGUUUU-- ..(((((((((......))).))))))..........((((.((((((((.......(.(((....))))((((..(((.((((.......))))))).))))..)))))))))))).-- ( -33.80) >DroSim_CAF1 62242 118 - 1 GUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGGAUGCCGCAAUUUAAGCGGAUUAAGACAAUGGUCAACGUUUU-- ..(((((((((......))).))))))..........((((.((((((((.......(.(((....))))((((..(((.((((.......))))))).))))..)))))))))))).-- ( -33.80) >DroEre_CAF1 55405 120 - 1 GUCAUAACCGAAUUAUAUCGCGUUAUGUUCGAACGCUGAACUGUUGGCCGAGUCGUUGACACAUUCGUGCGUCUAGAAUGCCACAAUUUGGCCGGAUUAAGACAAUGGUCAACGUUUCAG ..(((((((((......))).))))))........(((((.(((((((((.(((...(((.((....)).)))......((((.....))))........)))..))))))))).))))) ( -35.00) >DroYak_CAF1 58047 118 - 1 GUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCUGAGUCGUUGACACUUUCGUGCGUCUAGAAUGCCACAAUUGAAGGGGAUUAAGGCAAUGGUCAACGUUUG-- ..(((((((((......))).))))))..((((((.(((.((((((.((.(((((....).(((((((((((.....))).)))....))))).)))).)).))))))))).))))))-- ( -31.80) >consensus GUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGAAUGCCGCAAUUUAAGCGGAUUAAGACAAUGGUCAACGUUUU__ ..(((((((((......))).))))))..........((((.((((((((.......(.(((....))))((((..(((.((((.......))))))).))))..))))))))))))... (-30.26 = -30.06 + -0.20)

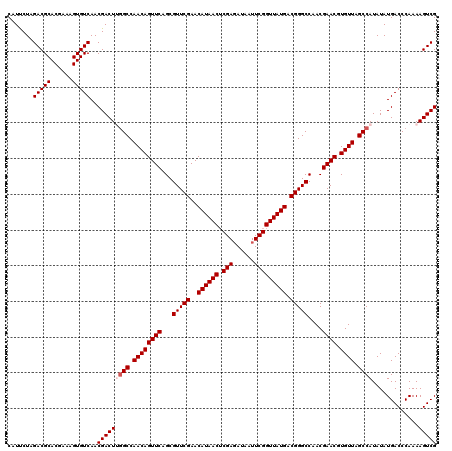
| Location | 4,071,419 – 4,071,539 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -32.93 |
| Energy contribution | -33.53 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
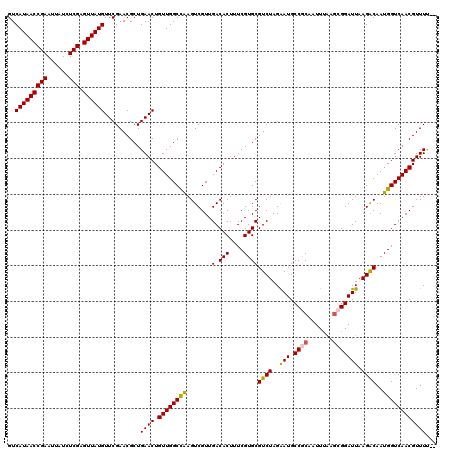
>2R_DroMel_CAF1 4071419 120 + 20766785 CAUUCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGACGGGCCAACGAACGUGUUAGCCAUAUAUGACCCAAAAGUCG .......(((((((....))))........((((.((((((((...(((((..((((((.((((....)))))))))).)))))....)))).)))).))))..............))). ( -35.60) >DroSec_CAF1 54101 120 + 1 CAUCCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGACGGGCCAACGAACGUGUUAGCCAUAUAUGACCCAAAAGUCG .......(((((((....))))........((((.((((((((...(((((..((((((.((((....)))))))))).)))))....)))).)))).))))..............))). ( -35.60) >DroSim_CAF1 62280 120 + 1 CAUCCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGACGGGCCAACGAACGUGUUAGCCAUAUAUGACCCAAAAGUCG .......(((((((....))))........((((.((((((((...(((((..((((((.((((....)))))))))).)))))....)))).)))).))))..............))). ( -35.60) >DroEre_CAF1 55445 120 + 1 CAUUCUAGACGCACGAAUGUGUCAACGACUCGGCCAACAGUUCAGCGUUCGAACAUAACGCGAUAUAAUUCGGUUAUGACGGGCCAACGAACGUGUUAGCCAUAUAUGACCCAAAAGUCG .......((((((....))))))........(((.((((((((...(((((..((((((.(((......))))))))).)))))....)))).)))).)))......(((......))). ( -34.90) >DroYak_CAF1 58085 120 + 1 CAUUCUAGACGCACGAAAGUGUCAACGACUCAGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGACGGGCCAACGAACGUGUUAGCCAUAUAUGACCCAAAAGUCG .......(((((((....))))..........((.((((((((...(((((..((((((.((((....)))))))))).)))))....)))).)))).))................))). ( -30.90) >consensus CAUUCUAGACGCACGAAAGUGUCAACGACUUGGCCAACAGUUCAGCGUUCGAACAUAACUCGAGAUAAUUCGGUUAUGACGGGCCAACGAACGUGUUAGCCAUAUAUGACCCAAAAGUCG .......(((((......)))))..(((((((((.((((((((...(((((..((((((.(((......))))))))).)))))....)))).)))).)))).............))))) (-32.93 = -33.53 + 0.60)

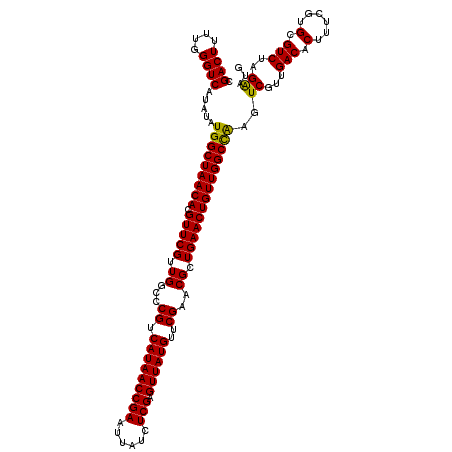
| Location | 4,071,419 – 4,071,539 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -36.64 |
| Energy contribution | -36.00 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
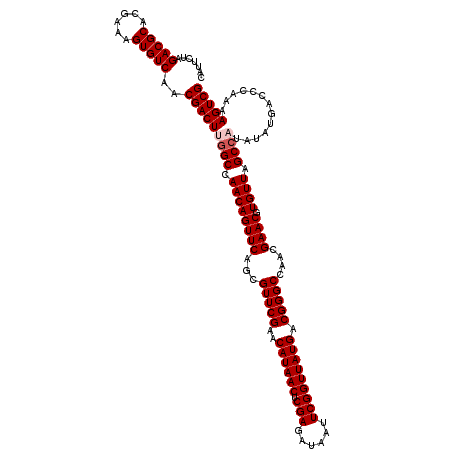
>2R_DroMel_CAF1 4071419 120 - 20766785 CGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGAAUG .((((....)))).....(((((((((.(((((.((...((.(((((((((......))).))))))..))..)).))))))))))))))..((...(((.(......).)))..))... ( -37.30) >DroSec_CAF1 54101 120 - 1 CGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGGAUG .((((....)))).....(((((((((.(((((.((...((.(((((((((......))).))))))..))..)).)))))))))))))).(((.(.(((.(......).))).).))). ( -39.10) >DroSim_CAF1 62280 120 - 1 CGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGGAUG .((((....)))).....(((((((((.(((((.((...((.(((((((((......))).))))))..))..)).)))))))))))))).(((.(.(((.(......).))).).))). ( -39.10) >DroEre_CAF1 55445 120 - 1 CGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUAUCGCGUUAUGUUCGAACGCUGAACUGUUGGCCGAGUCGUUGACACAUUCGUGCGUCUAGAAUG .((((....)))).....(((((((((.(((((.((...((.(((((((((......))).))))))..))..)).))))))))))))))..((...(((.((....)).)))..))... ( -36.90) >DroYak_CAF1 58085 120 - 1 CGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCUGAGUCGUUGACACUUUCGUGCGUCUAGAAUG .((((....))))......(((((((......)))))))((.(((((((((......))).))))))..)).((((((((.(((..((......))..)))..)))).))))........ ( -36.00) >consensus CGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAACUGUUGGCCAAGUCGUUGACACUUUCGUGCGUCUAGAAUG .((((....)))).....(((((((((.(((((.((...((.(((((((((......))).))))))..))..)).))))))))))))))..((...(((.(......).)))..))... (-36.64 = -36.00 + -0.64)

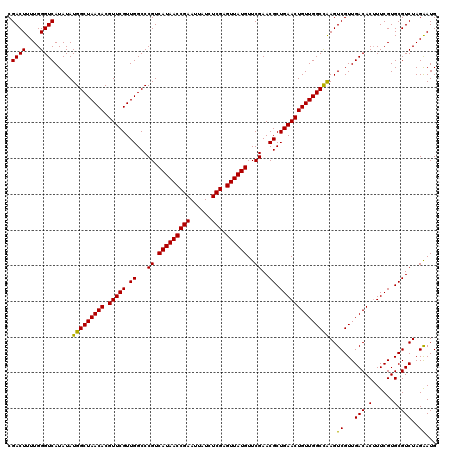
| Location | 4,071,459 – 4,071,579 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071459 120 - 20766785 CACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAA ................((((......)))).....((((((((.....((((((...(((.......)))....))))))..(((((((((......))).)))))).)))))))).... ( -34.30) >DroSec_CAF1 54141 120 - 1 CACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAA ................((((......)))).....((((((((.....((((((...(((.......)))....))))))..(((((((((......))).)))))).)))))))).... ( -34.30) >DroSim_CAF1 62320 120 - 1 CACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAA ................((((......)))).....((((((((.....((((((...(((.......)))....))))))..(((((((((......))).)))))).)))))))).... ( -34.30) >DroEre_CAF1 55485 120 - 1 CACAGAAAAAGUUAUCUGGCAUUGACUCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUAUCGCGUUAUGUUCGAACGCUGAA ..((((........))))((..((((.(((((((((....))).)))))))))).....(((((((......)))))))((.(((((((((......))).))))))..))...)).... ( -34.10) >DroYak_CAF1 58125 120 - 1 CGCAGAGAAAGUUAUCUGGCAUUGACUCCGAAAUUGUAUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAA ..((((........))))((..((((.(((((((((....))).)))))))))).....(((((((......)))))))((.(((((((((......))).))))))..))...)).... ( -34.80) >consensus CACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCCGUCAUAACCGAAUUAUCUCGAGUUAUGUUCGAACGCUGAA ..((((........))))((..((((.(((((((((....))).)))))))))).....(((((((......)))))))((.(((((((((......))).))))))..))...)).... (-34.36 = -34.36 + -0.00)

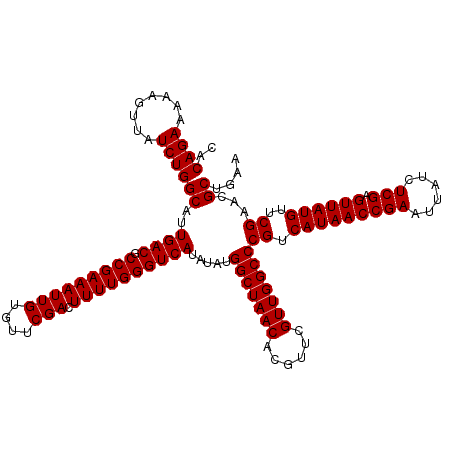
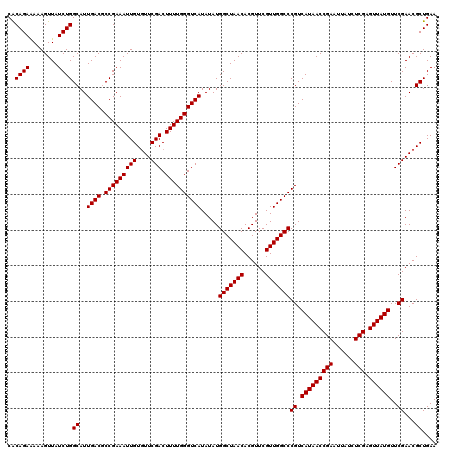
| Location | 4,071,499 – 4,071,619 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -33.40 |
| Energy contribution | -33.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071499 120 - 20766785 CUGUGGACAAUUAUCGUAUAUUUGCGCUGUCCAUCUGUGUCACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCC ..(((((((.....((((....)))).)))))))..((((..((((........))))))))((((.(((((((((....))).)))))))))).....(((((((......))))))). ( -33.80) >DroSec_CAF1 54181 120 - 1 CUGUGGACAAUUAUCGUAUAUUUGCGCUGCCCAUCUGUGUCACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCC .((((.........((((....))))..((((.((((.....))))(((((((....(((......)))(((......))))))))))))))))))...(((((((......))))))). ( -32.10) >DroSim_CAF1 62360 120 - 1 CUGUGGACAAUUAUCGUAUAUUUGCGCUGUCCAUCUGUGUCACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCC ..(((((((.....((((....)))).)))))))..((((..((((........))))))))((((.(((((((((....))).)))))))))).....(((((((......))))))). ( -33.80) >DroEre_CAF1 55525 120 - 1 CUGUGGACAAUUAUCGUAUAUUUGCGCUGUCCAUCUGUGUCACAGAAAAAGUUAUCUGGCAUUGACUCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCC ..(((((((.....((((....)))).)))))))..((((..((((........))))))))((((.(((((((((....))).)))))))))).....(((((((......))))))). ( -33.80) >DroYak_CAF1 58165 120 - 1 CUGUGGACAAUUAUCGCAUAUUUGCGCUGUCCAUCUGUGUCGCAGAGAAAGUUAUCUGGCAUUGACUCCGAAAUUGUAUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCC ..(((((((.....((((....)))).)))))))..((((..((((........))))))))((((.(((((((((....))).)))))))))).....(((((((......))))))). ( -36.60) >consensus CUGUGGACAAUUAUCGUAUAUUUGCGCUGUCCAUCUGUGUCACAGAAAAAGUUAUCUGGCAUUGACGCCGAAAUUGUGUUCGACUUUUGGGUCAUAUAUGGCUAACACGUUCGUUGGCCC ..(((((((.....((((....)))).)))))))..((((..((((........))))))))((((.(((((((((....))).)))))))))).....(((((((......))))))). (-33.40 = -33.44 + 0.04)

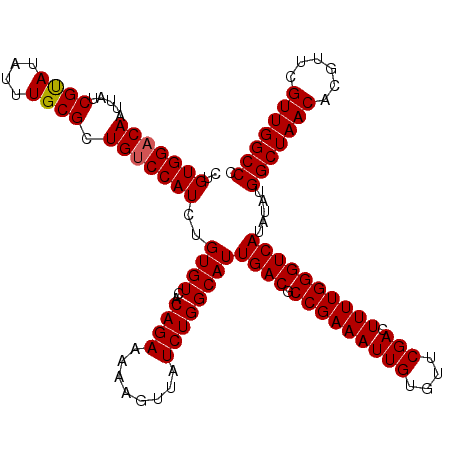
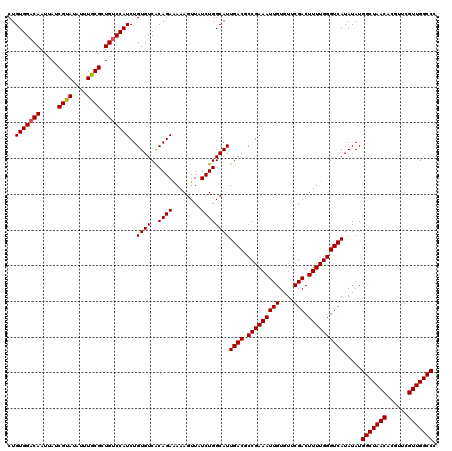
| Location | 4,071,619 – 4,071,739 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -20.82 |
| Energy contribution | -24.06 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071619 120 + 20766785 ACAUUGCAAGCGACCGAAUAAAACGUCAACUCAAUUUGAGUCCUCAAGUGCCAAGUGCCAAGGGCCAAGGGCCAAGUUGGGAUCAGGUUAAUCGGCGAAAGGUUACCGCCUUGCAAUGGA .(((((((((((.((((.(((........(((((((((.(((((...(.(((..........)))).))))))))))))))......))).))))(....).....)).))))))))).. ( -40.64) >DroSec_CAF1 54301 106 + 1 ACAUUGCAAGCGACCGAAUAAAACGUCAACUCAAUUUGAGUCCUCAAGUGCC--------------AAGGGCCAAGUUGGGAUCAGGUUAAUCGCCGAAAGGUUACCGCCUUGCAAUGGG .((((((((((((((((........))..(((((((((.(((((........--------------.))))))))))))))....))).....(((....)))...)).))))))))).. ( -38.40) >DroSim_CAF1 62480 106 + 1 ACAUUGCAAGCGACCGAAUAAAACGUCAACUCAAUUUGAGUCCUCAAGUGCC--------------AAGGGCCAAGUUGGGAUCAGGUUAAUCGCCGAAAGGUUACCGCCUUGCAAUGGG .((((((((((((((((........))..(((((((((.(((((........--------------.))))))))))))))....))).....(((....)))...)).))))))))).. ( -38.40) >DroEre_CAF1 55645 104 + 1 ACAUUGCCAGCGACAGAAUUAAACGUCAACUCAAUUUGAGUAGACAAGU--U--------------AAGGGCCAUGUCGGGGUCACGCUAAUCGGCGAAAAGUUACCGCCUUGCCAUUUA .....((.((((............(((.((((.....)))).)))....--.--------------...((((.......)))).))))....((((.........))))..))...... ( -24.40) >DroYak_CAF1 58285 106 + 1 ACAUUGCAAGCGACAGCAUAAAAUGUCAGCACAACUUGAGUAAACAAGUGCC--------------ACGGGCCAAGUCGGGAUCGGGUUAAUCGCCGAAAAGUUACCGCCUUGCAAUUGA ..((((((((.((((........)))).((...(((((......)))))...--------------..((......(((((((.......))).)))).......))))))))))))... ( -27.02) >consensus ACAUUGCAAGCGACCGAAUAAAACGUCAACUCAAUUUGAGUCCUCAAGUGCC______________AAGGGCCAAGUUGGGAUCAGGUUAAUCGCCGAAAGGUUACCGCCUUGCAAUGGA .(((((((((((((..........)))..(((((((((.(((((.......................))))))))))))))....((.(((((.......)))))))).))))))))).. (-20.82 = -24.06 + 3.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:07 2006