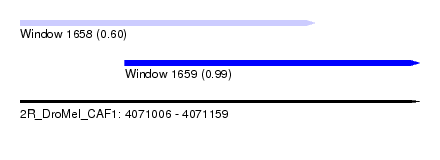
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,071,006 – 4,071,159 |
| Length | 153 |
| Max. P | 0.994876 |

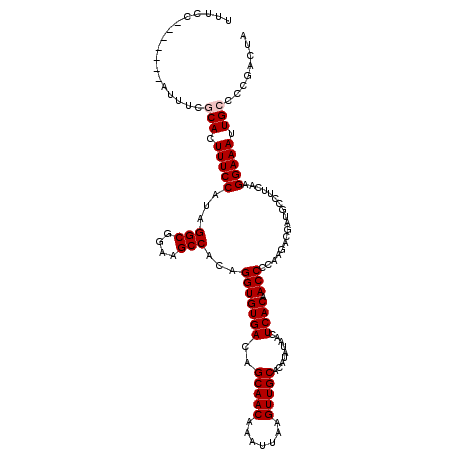
| Location | 4,071,006 – 4,071,119 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -23.01 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071006 113 + 20766785 UCGCUCAUAAAUUCAUGUCAAACAGGGCCAAGGUUGCCACUUUCC-------AUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCA ...............(((((.((..(((.(((((....)))))..-------.((((((.((......)))))))))))....)).)))))(((((.......)))))............ ( -25.60) >DroSec_CAF1 53684 113 + 1 GCGCUCAUAAAUUCAUGUCAAACAGGGCCAAGGUUGACACUUUCC-------AUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCA ...............((((((.(........).))))))......-------...((((((((......(((....)))..))))))))..(((((.......)))))............ ( -25.70) >DroSim_CAF1 61863 113 + 1 UCGCUCAUAAAUUCAUGUCAAACAGGGCCAAGGUUGCCACUUUCC-------GUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCA ...............(((((.((..(((.(((((....)))))..-------.((((((.((......)))))))))))....)).)))))(((((.......)))))............ ( -25.50) >DroEre_CAF1 55035 106 + 1 UCGCUCAUAAAUUCAUGUCAAACAGGGCCAAGGUUGCC--------------ACUUUCCACUUUCCCUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCA ...............(((((.((((((..(((((....--------------.......))))))))).(((....)))....)).)))))(((((.......)))))............ ( -26.50) >DroYak_CAF1 57654 120 + 1 UUGCUCAUAAAUUCAUGUCAAACAGGGCCAAGGUUGCCACUUGCCACUUGCCACUUUCCACUUUCCAUUGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCA ...............(((((.....(((..((((.((.....)).)))))))......(((((.....((((....)))).))))))))))(((((.......)))))............ ( -29.80) >consensus UCGCUCAUAAAUUCAUGUCAAACAGGGCCAAGGUUGCCACUUUCC_______AUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCA ..((((..................))))...(((((...................((((((((......(((....)))..))))))))..(((((.......))))).....))))).. (-23.01 = -23.17 + 0.16)

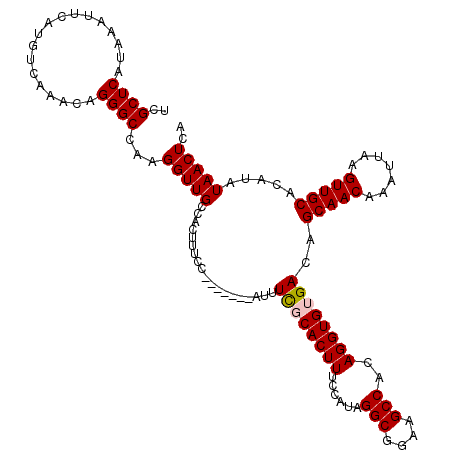
| Location | 4,071,046 – 4,071,159 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.07 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -24.68 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4071046 113 + 20766785 UUUCC-------AUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGAUGCCUUCAAGGAAAUUGCCCCGACUA .....-------.....(((.(((((...(((....)))...(((((((..(((((.......))))).........)))).)))..................))))).)))........ ( -27.30) >DroSec_CAF1 53724 113 + 1 UUUCC-------AUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGUCGAUGCCUUCAAGGAAAUUGCCCCGACUA .....-------.....(((.(((((...(((....)))..(((((((((.(((((.......))))).....(........)......))).))))))....))))).)))........ ( -28.30) >DroSim_CAF1 61903 113 + 1 UUUCC-------GUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGAUGCCUUCAAGGAAAUUGCCCCGACUA .....-------(((..(((.(((((...(((....)))...(((((((..(((((.......))))).........)))).)))..................))))).)))...))).. ( -28.20) >DroEre_CAF1 55073 108 + 1 ------------ACUUUCCACUUUCCCUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGACGCCUUCAAGGAAAUUGCCCAGAAUA ------------.((...((.(((((...(((....)))..((((((....(((((.......)))))............(........)...))))))....))))).))...)).... ( -24.50) >DroYak_CAF1 57694 119 + 1 UUGCCACUUGCCACUUUCCACUUUCCAUUGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACAGGGCCUUCAAGGAAAUUGCCCGG-CUA ..(((.((((........(((...((..((((....))))..)).)))...(((((.......)))))..................))))...((((.(((...)))...))))))-).. ( -34.10) >consensus UUUCC_______AUUUCGCACUUUCCAUAGGCGGAAGCCACAGGUGUGACAGCAACAAAUUAAGUUGCACAUAUAACUCACAACCCCAAGACGAUGCCUUCAAGGAAAUUGCCCCGACUA .................(((.(((((...(((....)))...(((((((..(((((.......))))).........)))).)))..................))))).)))........ (-24.68 = -25.08 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:57 2006