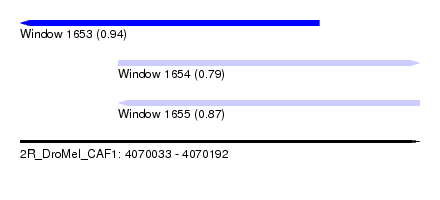
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,070,033 – 4,070,192 |
| Length | 159 |
| Max. P | 0.943399 |

| Location | 4,070,033 – 4,070,152 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4070033 119 - 20766785 UUGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGCUUGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAAAAGUGCAACAA-AAAGAAUUCGAGAUAGAAAUCAAUUUUU ....((((..(.....(((((((.....)))))))((((((((((..(((....)))...)).))))))))...........)..))))..-...((.(((......))).))....... ( -34.30) >DroSec_CAF1 52686 118 - 1 CCGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGC-UGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAAAACUGCAACAA-AAAGAAUUUGAGAUAGAAAUCAAUUUUU .(((((((.((...(((((((((.....)))))))))(-((((((..(((....)))...)).))))))))))))))..............-..((((.((((........)))).)))) ( -32.50) >DroSim_CAF1 60864 118 - 1 UCGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGC-UGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAAAACUGCAACAA-AAAGAAUUCGAGAUAGAAAUCAAUUUUU ((((((((.((...(((((((((.....)))))))))(-((((((..(((....)))...)).))))))))))))))).............-...((.(((......))).))....... ( -34.20) >DroEre_CAF1 54058 120 - 1 UUGCGUUGCGCAACUUCAGUUUAAAAACUAAACGGGGCUUGCUGUCUGCCUCUGGGCAAAACAAUUAGGCCAACGCGGAAAAAUGCACCAAAAAAGUAUCCGGGAUAGAAAUCAAUUUUU ..(((((((((((((((.(((((.....))))))))).))))(((.((((....))))..))).....).))))))(((....(((.........))))))..(((....)))....... ( -26.30) >DroYak_CAF1 56717 118 - 1 UUGCGUUGUGCAACUUCAGUUCAAAAACUGAACGGGGCGGGCUGUCUGCCUUUGGGCA-AACAAUUAGGCCAACGCGGAAAAGUGCAACAA-AAAGUAUCCGAGAUAGCAAUCAAUUUUU ((((((((..(...(((((((....)))))))((.(...((((((.((((....))))-.)).....))))..).)).....)..))))..-....((((...))))))))......... ( -33.70) >consensus UUGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGC_UGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAAAACUGCAACAA_AAAGAAUUCGAGAUAGAAAUCAAUUUUU ((((((((.((...(((((((((.....))))))))).....(((..(((....)))...))).....)))))))))).........................(((....)))....... (-25.16 = -25.04 + -0.12)

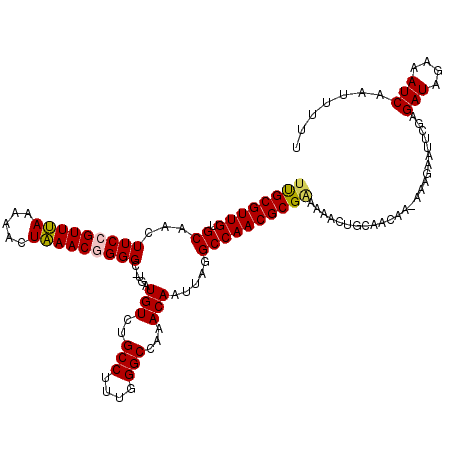
| Location | 4,070,072 – 4,070,192 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4070072 120 + 20766785 UUUCGCGUUGGCCUAAUUGUUUGGCCCAAAGGCAGACAUCAAGCCCCGUUUAGUUUUUAAACGGAAGUUGCACAACGCAAGAAAUAAAAGGCUGAAAGGAUUUAGAACAUGGCAAAAAGU ....((..((..((((.((((((.((....))))))))(((.(((((((((((...))))))))...((((.....)))).........))))))......))))..))..))....... ( -29.60) >DroSec_CAF1 52725 119 + 1 UUUCGCGUUGGCCUAAUUGUUUGGCCCAAAGGCAGACAUCA-GCCCCGUUUAGUUUUUAAACGGAAGUUGCACAACGCGGAAAACAAAAGGCUGAAAGGAUUUUGAACAUGGCAAAAAGU ((((((((((.......((((((.((....)))))))).((-((.((((((((...))))))))..))))..))))))))))..(((((..((....)).)))))............... ( -34.80) >DroSim_CAF1 60903 119 + 1 UUUCGCGUUGGCCUAAUUGUUUGGCCCAAAGGCAGACAUCA-GCCCCGUUUAGUUUUUAAACGGAAGUUGCACAACGCGAAAAACAAAAGGCUGAAAGGAUUUUGAACAUGGCAAAAAGU ((((((((((.......((((((.((....)))))))).((-((.((((((((...))))))))..))))..))))))))))..(((((..((....)).)))))............... ( -35.30) >DroEre_CAF1 54098 119 + 1 UUCCGCGUUGGCCUAAUUGUUUUGCCCAGAGGCAGACAGCAAGCCCCGUUUAGUUUUUAAACUGAAGUUGCGCAACGCAAAAAC-CAAAGGCUGAAAGGAUUUUCAACAUGGCAAAAAGU .(((...(..((((..((((((((((....))))))..))))......(((((((....))))))).(((((...)))))....-...))))..)..))).................... ( -28.60) >DroYak_CAF1 56756 119 + 1 UUCCGCGUUGGCCUAAUUGUU-UGCCCAAAGGCAGACAGCCCGCCCCGUUCAGUUUUUGAACUGAAGUUGCACAACGCAAAAAACAAAACGCUGAAAGGAUUUUGAAUAUGGCAAAAAGU ..((((((((((....(((((-((((....))))))))).........(((((((....)))))))...)).))))))......(((((..((....)).))))).....))........ ( -33.60) >consensus UUUCGCGUUGGCCUAAUUGUUUGGCCCAAAGGCAGACAUCA_GCCCCGUUUAGUUUUUAAACGGAAGUUGCACAACGCAAAAAACAAAAGGCUGAAAGGAUUUUGAACAUGGCAAAAAGU ....((...(((.....(((((.(((....))))))))....)))((.((((((((((.(((....)))((.....))........)))))))))).))............))....... (-25.98 = -26.22 + 0.24)

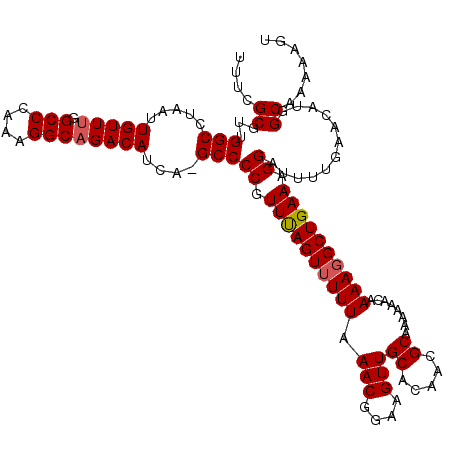
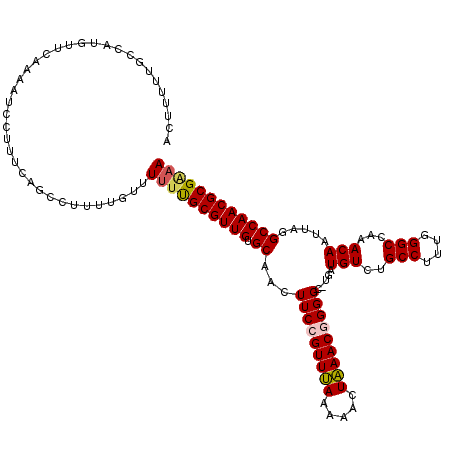
| Location | 4,070,072 – 4,070,192 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.76 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4070072 120 - 20766785 ACUUUUUGCCAUGUUCUAAAUCCUUUCAGCCUUUUAUUUCUUGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGCUUGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAA ........................................(((((((.........(((((((.....)))))))((((((((((..(((....)))...)).))))))))))))))).. ( -30.10) >DroSec_CAF1 52725 119 - 1 ACUUUUUGCCAUGUUCAAAAUCCUUUCAGCCUUUUGUUUUCCGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGC-UGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAA .........................................(((((((.((...(((((((((.....)))))))))(-((((((..(((....)))...)).))))))))))))))... ( -30.20) >DroSim_CAF1 60903 119 - 1 ACUUUUUGCCAUGUUCAAAAUCCUUUCAGCCUUUUGUUUUUCGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGC-UGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAA ......................................((((((((((.((...(((((((((.....)))))))))(-((((((..(((....)))...)).))))))))))))))))) ( -32.70) >DroEre_CAF1 54098 119 - 1 ACUUUUUGCCAUGUUGAAAAUCCUUUCAGCCUUUG-GUUUUUGCGUUGCGCAACUUCAGUUUAAAAACUAAACGGGGCUUGCUGUCUGCCUCUGGGCAAAACAAUUAGGCCAACGCGGAA .......((((.(((((((....)))))))...))-))(((((((((((((((((((.(((((.....))))))))).))))(((.((((....))))..))).....).)))))))))) ( -35.90) >DroYak_CAF1 56756 119 - 1 ACUUUUUGCCAUAUUCAAAAUCCUUUCAGCGUUUUGUUUUUUGCGUUGUGCAACUUCAGUUCAAAAACUGAACGGGGCGGGCUGUCUGCCUUUGGGCA-AACAAUUAGGCCAACGCGGAA ...............((((((.(.....).))))))..(((((((((.(((..((((((((....))))))).)..)))((((((.((((....))))-.)).....))))))))))))) ( -32.40) >consensus ACUUUUUGCCAUGUUCAAAAUCCUUUCAGCCUUUUGUUUUUUGCGUUGUGCAACUUCCGUUUAAAAACUAAACGGGGC_UGAUGUCUGCCUUUGGGCCAAACAAUUAGGCCAACGCGAAA ......................................((((((((((.((...(((((((((.....))))))))).....(((..(((....)))...))).....)))))))))))) (-25.68 = -25.76 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:53 2006