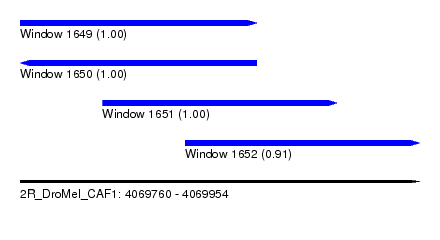
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,069,760 – 4,069,954 |
| Length | 194 |
| Max. P | 0.999979 |

| Location | 4,069,760 – 4,069,875 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.96 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -31.98 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4069760 115 + 20766785 UGGUCAACUUUUAUGGGCAGCCAAUAACGCCAGGGGAUUUGGCAAUUCUCUUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA----- ((((((.(.(((((((((((((((....(((((.....)))))................(((...........))).......))))))...)))))))))..).))))))....----- ( -36.00) >DroSec_CAF1 52414 115 + 1 UGGCCAACUUUUAUGGGUAGCCAAUAACGCCAGGGGAUUUGGCAGUUCUCCUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA----- ((((((.(.(((((((((((((((.......((((((.........)))))).......(((...........))).......))))))...)))))))))..).))))))....----- ( -37.50) >DroSim_CAF1 60591 115 + 1 UGGCCAACUUUUAUGGGUAGCCAAUAACGCCAGGGGAUUUGGCAAUUCUCCUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA----- ((((((.(.(((((((((((((((.......((((((.........)))))).......(((...........))).......))))))...)))))))))..).))))))....----- ( -37.50) >DroEre_CAF1 53780 119 + 1 UGGCCAACUUUUAUGGUCAGCCAAUAACCCCAGGGGAUUCGGCAAUUCUCCUCUUC-CCGCUUCGAAAUCAAUAGCAAUAAUUCUGGCUAGCGCCCAUAAAUUGCUGGCCACAUAUAUAC ((((((.(.(((((((.((((((..........((((...((.......))...))-))(((...........)))........)))))...).)))))))..).))))))......... ( -31.60) >DroYak_CAF1 56448 112 + 1 GGGCCAACUUUUAUGGUCAGCCAAUAACCCUAGGGGAUUUGGCAAUU--CCUCUUC-UCGCUUCUAAAUCAAUAGCAAUAAUUCUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA----- .(((((.......))))).(((((...((....))...)))))....--.......-..(((...........)))........(((((((((.........)))))))))....----- ( -31.20) >consensus UGGCCAACUUUUAUGGGCAGCCAAUAACGCCAGGGGAUUUGGCAAUUCUCCUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA_____ .(((((.(.((((((((((((((........((((((.........)))))).......(((...........)))........)))))...)))))))))..).))))).......... (-31.98 = -32.22 + 0.24)

| Location | 4,069,760 – 4,069,875 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.96 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -32.42 |
| Energy contribution | -32.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4069760 115 - 20766785 -----UUUGUGGCCAGCAAUUUAUGGGCGCUAGCCAAAAUUAUUGCUAUUGAUUUAGAAGCGAAGAAGAAGAGAAUUGCCAAAUCCCCUGGCGUUAUUGGCUGCCCAUAAAAGUUGACCA -----....(((.((((..(((((((((...((((((.....(((((...........)))))..............((((.......))))....))))))))))))))).)))).))) ( -34.90) >DroSec_CAF1 52414 115 - 1 -----UUUGUGGCCAGCAAUUUAUGGGCGCUAGCCAAAAUUAUUGCUAUUGAUUUAGAAGCGAAGAAGAGGAGAACUGCCAAAUCCCCUGGCGUUAUUGGCUACCCAUAAAAGUUGGCCA -----....((((((((..((((((((...(((((((.....(((((...........)))))..........(((.((((.......))))))).))))))))))))))).)))))))) ( -40.50) >DroSim_CAF1 60591 115 - 1 -----UUUGUGGCCAGCAAUUUAUGGGCGCUAGCCAAAAUUAUUGCUAUUGAUUUAGAAGCGAAGAAGAGGAGAAUUGCCAAAUCCCCUGGCGUUAUUGGCUACCCAUAAAAGUUGGCCA -----....((((((((..((((((((...(((((((.....(((((...........)))))..............((((.......))))....))))))))))))))).)))))))) ( -39.90) >DroEre_CAF1 53780 119 - 1 GUAUAUAUGUGGCCAGCAAUUUAUGGGCGCUAGCCAGAAUUAUUGCUAUUGAUUUCGAAGCGG-GAAGAGGAGAAUUGCCGAAUCCCCUGGGGUUAUUGGCUGACCAUAAAAGUUGGCCA .........((((((((..(((((((((....))).........((((.((((..(..((.((-((...((.......))...)))))))..)))).))))....)))))).)))))))) ( -40.20) >DroYak_CAF1 56448 112 - 1 -----UUUGUGGCCAGCAAUUUAUGGGCGCUAGCCAGAAUUAUUGCUAUUGAUUUAGAAGCGA-GAAGAGG--AAUUGCCAAAUCCCCUAGGGUUAUUGGCUGACCAUAAAAGUUGGCCC -----.....(((((((..(((((((.((((...(.((((((.......)))))).).)))).-.......--....(((((..(((...)))...)))))...))))))).))))))). ( -34.60) >consensus _____UUUGUGGCCAGCAAUUUAUGGGCGCUAGCCAAAAUUAUUGCUAUUGAUUUAGAAGCGAAGAAGAGGAGAAUUGCCAAAUCCCCUGGCGUUAUUGGCUGCCCAUAAAAGUUGGCCA ..........(((((((..((((((((...(((((((.....(((((...........))))).............(((((.......)))))...))))))))))))))).))))))). (-32.42 = -32.82 + 0.40)

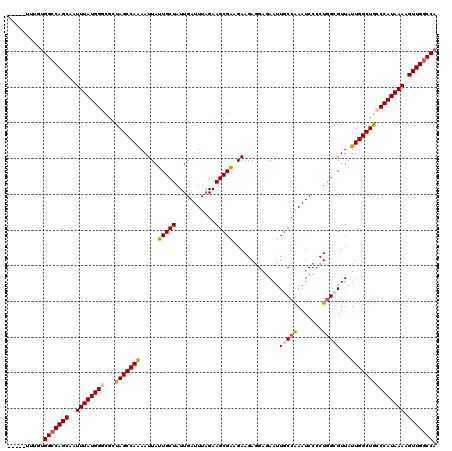
| Location | 4,069,800 – 4,069,914 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
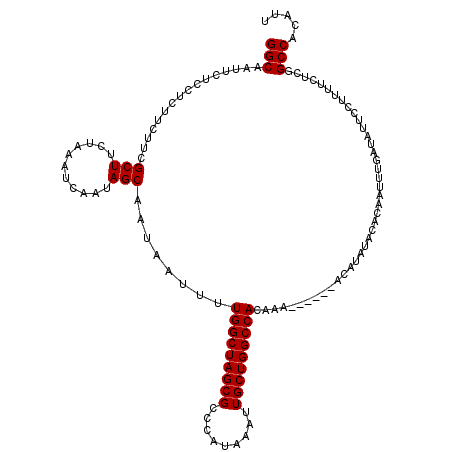
>2R_DroMel_CAF1 4069800 114 + 20766785 GGCAAUUCUCUUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------ACAUAUACACAAUUUGAUAUUCCUUUUCUCGGCCACAUU (((................(((...........)))........(((((((((.........)))))))))....------...............................)))..... ( -20.00) >DroSec_CAF1 52454 114 + 1 GGCAGUUCUCCUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------ACAUAUACACAAUUUGAUGUUCCUUUUCUCGGCCACAUU (((................(((...........)))........(((((((((.........)))))))))....------...............................)))..... ( -20.00) >DroSim_CAF1 60631 114 + 1 GGCAAUUCUCCUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------ACAUAUACACAAUUUGAUAUUCCUUUUCUCGGCCACAUU (((................(((...........)))........(((((((((.........)))))))))....------...............................)))..... ( -20.00) >DroEre_CAF1 53820 119 + 1 GGCAAUUCUCCUCUUC-CCGCUUCGAAAUCAAUAGCAAUAAUUCUGGCUAGCGCCCAUAAAUUGCUGGCCACAUAUAUACCAUAUAUACAGAAUUUGAUAUUCCUUUUCUCAGCCACAUU (((.............-..(((...........)))...((((((((((((((.........))))))))..((((((...))))))..)))))).................)))..... ( -22.90) >DroYak_CAF1 56488 111 + 1 GGCAAUU--CCUCUUC-UCGCUUCUAAAUCAAUAGCAAUAAUUCUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------AUAUAUACAGAAUUUGAUAUUUCUUUUCUCAGCCGCAUU (((....--.......-..(((...........)))........(((((((((.........)))))))))....------........((((...((....))..))))..)))..... ( -21.30) >consensus GGCAAUUCUCCUCUUCUUCGCUUCUAAAUCAAUAGCAAUAAUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA______ACAUAUACACAAUUUGAUAUUCCUUUUCUCGGCCACAUU (((................(((...........)))........(((((((((.........))))))))).........................................)))..... (-20.04 = -20.04 + -0.00)

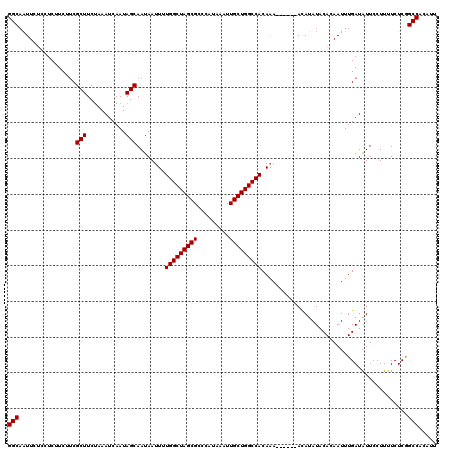
| Location | 4,069,840 – 4,069,954 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -18.71 |
| Energy contribution | -18.59 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4069840 114 + 20766785 AUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------ACAUAUACACAAUUUGAUAUUCCUUUUCUCGGCCACAUUAUCAGACCGCAUACUCACACACGUGUUGGCCAUAAAUCUA ....(((((((((.........)))))))))....------..............................((((((((.....((........))......))).)))))......... ( -22.00) >DroSec_CAF1 52494 113 + 1 AUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------ACAUAUACACAAUUUGAUGUUCCUUUUCUCGGCCACAUUAUCAGA-CGCAUACUCACACACGUGUUGGCCAUAAAUCUA ....((((((((((........((.(((((.....------......((((.....).)))..........)))))))......((-.......))......))))))))))........ ( -22.30) >DroSim_CAF1 60671 114 + 1 AUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------ACAUAUACACAAUUUGAUAUUCCUUUUCUCGGCCACAUUAUCAGACCGCAUACUCACACACGUGUUGGCCAUAAAUCUA ....(((((((((.........)))))))))....------..............................((((((((.....((........))......))).)))))......... ( -22.00) >DroEre_CAF1 53859 120 + 1 AUUCUGGCUAGCGCCCAUAAAUUGCUGGCCACAUAUAUACCAUAUAUACAGAAUUUGAUAUUCCUUUUCUCAGCCACAUUAUCAGACCGCAUACUCACACACGUGUUGGCCAUAAAUCUA ....((((((((((........(((.((....((((((...))))))......(((((((...................))))))))))))...........))))))))))........ ( -21.32) >DroYak_CAF1 56525 114 + 1 AUUCUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA------AUAUAUACAGAAUUUGAUAUUUCUUUUCUCAGCCGCAUUAUCAGACCGCAAACUCACACACGCGUUGACCAUAAAUCAA ....(((((((((.........)))))))))(((.------............(((((((...................))))))).(((............))))))............ ( -17.71) >consensus AUUUUGGCUAGCGCCCAUAAAUUGCUGGCCACAAA______ACAUAUACACAAUUUGAUAUUCCUUUUCUCGGCCACAUUAUCAGACCGCAUACUCACACACGUGUUGGCCAUAAAUCUA ....(((((((((((((........))).........................(((((((...................)))))))................))))))))))........ (-18.71 = -18.59 + -0.12)

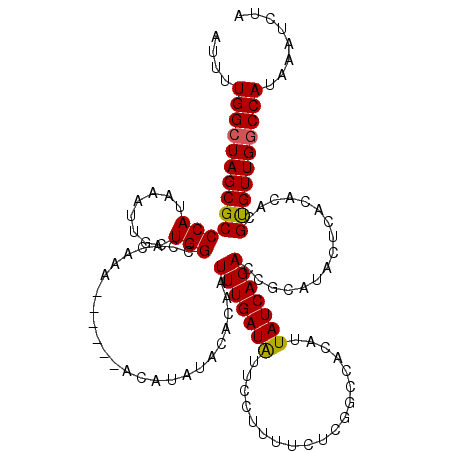
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:50 2006