
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
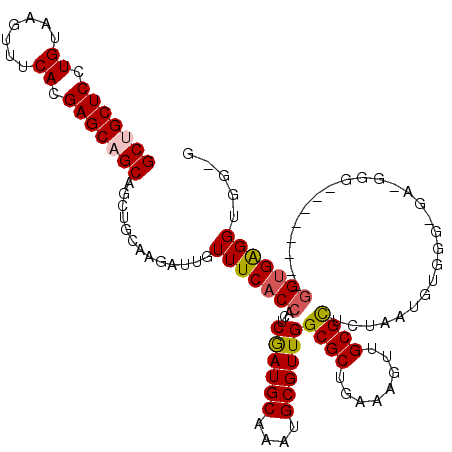
| Location | 4,068,738 – 4,068,853 |
| Length | 115 |
| Max. P | 0.718159 |

| Location | 4,068,738 – 4,068,853 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -30.88 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
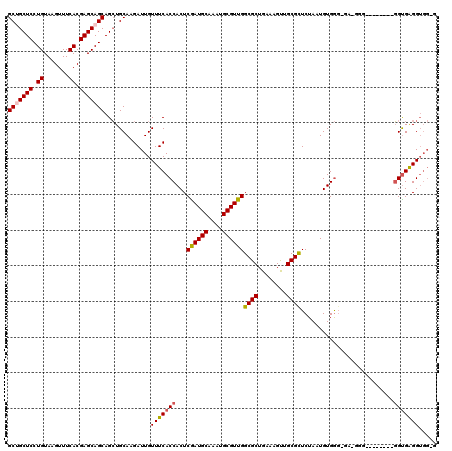
>2R_DroMel_CAF1 4068738 115 + 20766785 GCUGCUCCUGUAAGUUUCACGAGCAGCAGAUGCAAGAUUGUUUCACCACUCGAUGCAAAUGCGUUGGCGCUGAAAGCAGCGCUCUAAUGUGAGGGGGGGGGGG-----UGUGAGGUGCUG (((((((.((.......)).)))))))((.(((.(.(((.(..(.((.((((((((....)))))(((((((....))))))).......))).)))..).))-----).)...))))). ( -41.50) >DroSec_CAF1 51381 108 + 1 GCAGCUCCUGUAAGUUUCACGAGCAGCAGCUGCAAGAUUGUUUCACCACUCGAUGCAAAUGCGUUGGCGCUGAAAGUUGCGCUCUAAUGUGGGUGU------------GGUGAGGUGGAG ((((((.((((..(.....)..)))).))))))....(..(((((((((.((((((....))))))((((........))))............))------------)))))))..).. ( -44.10) >DroSim_CAF1 59572 106 + 1 GCAGCUCCUGUAAGUUUCACGAGCAGCAGCUGCAAGAUUGUUUCACCACUCGAUGCAAAUGCGUUGGCGCUGAAAGUUGCGCUCUAAUGUGGG--G------------GGUGAGGUGGGG ((((((.((((..(.....)..)))).))))))....(..(((((((.((((((((....)))))(((((........))))).......)))--.------------)))))))..).. ( -41.90) >DroEre_CAF1 52835 108 + 1 GCUGCUCCUGUAAGUUUCACGAGCAGCAGCUGCAAGAUUGUUUCACCACUCAAUGCAAAUGCGUUGGCGCUGAAAGUUGCGCUCUAAUGUGGC--AAGGG--------GGUGGGGUGG-- (((((((.((.......)).)))))))..........(..(..((((.(((..(((....((((((((((........)))))..))))).))--).)))--------))))..)..)-- ( -39.80) >DroYak_CAF1 55466 117 + 1 GCUGCUCCUGUAAGUUUCACGAGCAGCAGCUGCAAGAUUGUUUCACCACUCGAUGCAAAUGCGUUGGCGCUGAAAGUAGCGUUCUAGUGUGGGGGAAGGGCGGAGUAUGGCGGGGA-G-- (((((((.((.......)).)))))))..((((....((.(..(((.(((((((((....))))))((((((....))))))...))))))..).))..)))).............-.-- ( -40.00) >consensus GCUGCUCCUGUAAGUUUCACGAGCAGCAGCUGCAAGAUUGUUUCACCACUCGAUGCAAAUGCGUUGGCGCUGAAAGUUGCGCUCUAAUGUGGG_GA_GGG________GGUGAGGUGG_G (((((((.((.......)).))))))).............(((((((...((((((....))))))((((........))))..........................)))))))..... (-30.88 = -31.12 + 0.24)

| Location | 4,068,738 – 4,068,853 |
|---|---|
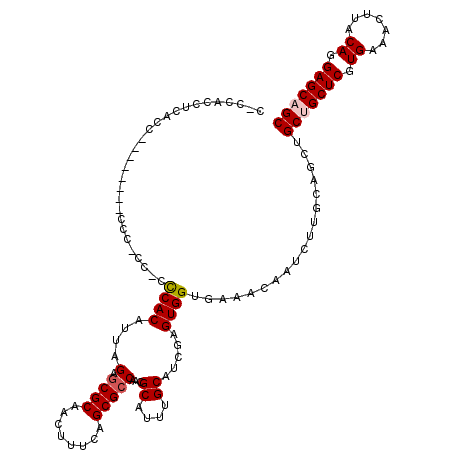
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -23.80 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
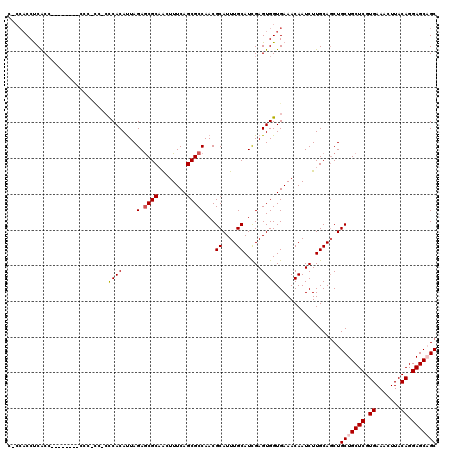
>2R_DroMel_CAF1 4068738 115 - 20766785 CAGCACCUCACA-----CCCCCCCCCCCUCACAUUAGAGCGCUGCUUUCAGCGCCAACGCAUUUGCAUCGAGUGGUGAAACAAUCUUGCAUCUGCUGCUCGUGAAACUUACAGGAGCAGC ..(((.......-----........(((((......(.((((((....)))))))...((....))...))).)).((.....)).)))....(((((((.((.......)).))))))) ( -32.00) >DroSec_CAF1 51381 108 - 1 CUCCACCUCACC------------ACACCCACAUUAGAGCGCAACUUUCAGCGCCAACGCAUUUGCAUCGAGUGGUGAAACAAUCUUGCAGCUGCUGCUCGUGAAACUUACAGGAGCUGC .......(((((------------((..........(.((((........)))))...((....)).....))))))).........((((((.(((.............))).)))))) ( -32.72) >DroSim_CAF1 59572 106 - 1 CCCCACCUCACC------------C--CCCACAUUAGAGCGCAACUUUCAGCGCCAACGCAUUUGCAUCGAGUGGUGAAACAAUCUUGCAGCUGCUGCUCGUGAAACUUACAGGAGCUGC .......(((((------------.--(.(......(.((((........)))))...((....))...).).))))).........((((((.(((.............))).)))))) ( -27.72) >DroEre_CAF1 52835 108 - 1 --CCACCCCACC--------CCCUU--GCCACAUUAGAGCGCAACUUUCAGCGCCAACGCAUUUGCAUUGAGUGGUGAAACAAUCUUGCAGCUGCUGCUCGUGAAACUUACAGGAGCAGC --..........--------...(.--.((((......((((........))))(((.((....)).))).))))..)...............(((((((.((.......)).))))))) ( -32.00) >DroYak_CAF1 55466 117 - 1 --C-UCCCCGCCAUACUCCGCCCUUCCCCCACACUAGAACGCUACUUUCAGCGCCAACGCAUUUGCAUCGAGUGGUGAAACAAUCUUGCAGCUGCUGCUCGUGAAACUUACAGGAGCAGC --.-....((((..((((.....................((((......)))).....((....))...))))))))................(((((((.((.......)).))))))) ( -27.60) >consensus C_CCACCUCACC________CCC_CC_CCCACAUUAGAGCGCAACUUUCAGCGCCAACGCAUUUGCAUCGAGUGGUGAAACAAUCUUGCAGCUGCUGCUCGUGAAACUUACAGGAGCAGC ............................((((....(.((((........)))))...((....)).....))))..................(((((((.((.......)).))))))) (-23.80 = -24.24 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:47 2006