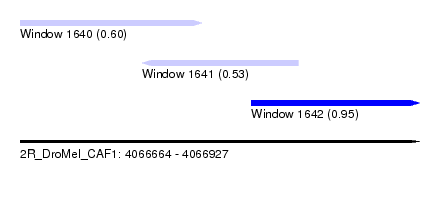
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,066,664 – 4,066,927 |
| Length | 263 |
| Max. P | 0.952118 |

| Location | 4,066,664 – 4,066,784 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -29.60 |
| Energy contribution | -30.04 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4066664 120 + 20766785 GACAUCGUCAAAUUAAAUGGCCUGGCCAGAAUGCAACAGUUAAAACGGCACACAACAAUGGCCAGAAGCGGGCACUUGAAGUCCGGCCAAAUGGCCUACUGGGCGCAGUCCGGAAAACUG (((...))).........(((((((((.....((....((....))(((.((......)))))....))((((.......)))))))))...))))..((((((...))))))....... ( -35.40) >DroSec_CAF1 49208 120 + 1 GACAUCGUCAAAUUAAAUGGCCUGGCCACAAUGCAACAGUUAAAACGGCAGACAACAAUGGCCAGAAGCGGGCACUUGAAGUCCGGCCAAAUGGCCUACUGGGCGCAGCCCGGAAAACUG (((...))).........(((((((((....(((....((....)).)))(((..(((((.((......)).)).)))..))).)))))...))))..((((((...))))))....... ( -37.90) >DroSim_CAF1 57448 120 + 1 GACAUCGUCAAAUUAAAUGGCCUGGCCACAAUGCAACAGUUAAAACGGCAGACAACAAUGGCCAGAAGCGGGCACUUGAAGUCCGGCCAAAUGGCCUACUGGGCGCAGCCCGGAAAACUG (((...))).........(((((((((....(((....((....)).)))(((..(((((.((......)).)).)))..))).)))))...))))..((((((...))))))....... ( -37.90) >DroEre_CAF1 50526 120 + 1 GCCAUCGUCAAACCCAAUGGCCUGGCCACAAUGCAACAGCUAAAACGGCGGAGAACAAUGGCCAGAAGCAGGCACUUGAAGUCCGGCCAAAUGGCCUACUGGCCGCAGUGCGGAAAACUG (((((.(.......).)))))....((.((.(((....(((.....)))..........((((((.....((.((.....))))((((....))))..))))))))).)).))....... ( -37.80) >DroYak_CAF1 53290 120 + 1 GACAUCGUCAAAUUAAAUGGCCUGGCCACAAUGCAACAGUUAAAACAGCGGACAACAAUGGCCAGAAGCAGGCACUUGAAGUCCGGCCAAAUGGCCUACUGGGCGCAGCACGGAAAACUG ....(((((........((((...))))...(((..((((.........((((..(((..(((.......)))..)))..))))((((....)))).))))...)))).))))....... ( -33.20) >consensus GACAUCGUCAAAUUAAAUGGCCUGGCCACAAUGCAACAGUUAAAACGGCAGACAACAAUGGCCAGAAGCGGGCACUUGAAGUCCGGCCAAAUGGCCUACUGGGCGCAGCCCGGAAAACUG (((.((((((.......))))(((((((...(((....(......).)))........)))))))............)).))).((((....))))..((((((...))))))....... (-29.60 = -30.04 + 0.44)

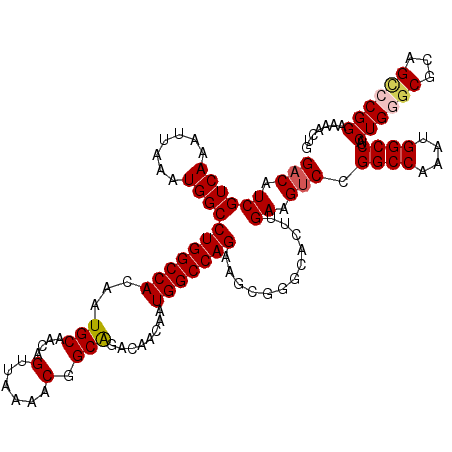
| Location | 4,066,744 – 4,066,847 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.75 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4066744 103 - 20766785 AGCUGUUCUUAAGGUUGCCCCCAC------UAACCGC----CCAUUCG-------CAUCCCCGUUGGGAUGCUGAUGUUGCAGUUUUCCGGACUGCGCCCAGUAGGCCAUUUGGCCGGAC .((((.......(((((.......------)))))((----.((((.(-------((((((....))))))).))))..((((((.....)))))))).)))).((((....)))).... ( -39.10) >DroSec_CAF1 49288 86 - 1 AGCUGUUCUUAAGUUUGCCCCC----------------------------------AUCCCCGUUGCGAUGCUGAUGUUGCAGUUUUCCGGGCUGCGCCCAGUAGGCCAUUUGGCCGGAC ((((((.....(((((((....----------------------------------.........)))).)))......)))))).(((((((...))))....((((....))))))). ( -25.92) >DroSim_CAF1 57528 86 - 1 AGCUGUUCUUAAGUUUGCCCCC----------------------------------CUCCCCGUUGCGAUGCUGAUGUUGCAGUUUUCCGGGCUGCGCCCAGUAGGCCAUUUGGCCGGAC ((((((.....(((((((....----------------------------------.........)))).)))......)))))).(((((((...))))....((((....))))))). ( -25.92) >DroEre_CAF1 50606 97 - 1 GUCUGUUCUCAAGGC----------------AUCCCCACCACCAUCCC-------CAUUCCCAUUCCGAUGCUGAUGUUGCAGUUUUCCGCACUGCGGCCAGUAGGCCAUUUGGCCGGAC ((((.......))))----------------.................-------.........(((..(((((..((((((((.......)))))))))))))((((....))))))). ( -28.60) >DroYak_CAF1 53370 120 - 1 GGCAUUUUAUAAGGCUUAACCCACCUGCCCUAUCCGCAUCCGCAUCCCCCCCCAGCAUCCCCAAUCCGCUGCUGAUGUUGCAGUUUUCCGUGCUGCGCCCAGUAGGCCAUUUGGCCGGAC .((.....(((.(((...........))).)))..)).(((((.........(((((.(........).))))).....(((((.......)))))))......((((....))))))). ( -29.50) >consensus AGCUGUUCUUAAGGUUGCCCCC_________A_CC_C_____CAU_C________CAUCCCCGUUGCGAUGCUGAUGUUGCAGUUUUCCGGGCUGCGCCCAGUAGGCCAUUUGGCCGGAC ............................................................((.......(((((..(..((((((.....))))))..))))))((((....)))))).. (-21.30 = -21.46 + 0.16)

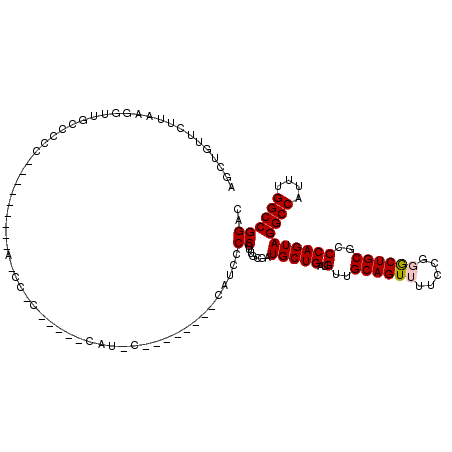
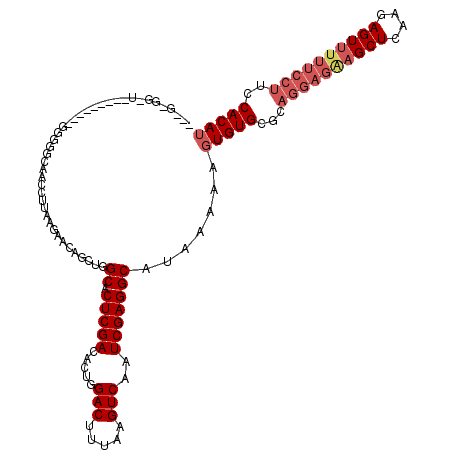
| Location | 4,066,816 – 4,066,927 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -22.86 |
| Energy contribution | -24.34 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4066816 111 + 20766785 ---GCGGUUA------GUGGGGGCAACCUUAAGAACAGCUGGCACUCGACACUGGACUUUAAGUCAAUCGAGGCAUAAAAAGUGUGCGCAGGAGAAGCUCAAGAGUUUUUCCUUCCACAU ---((.(((.------.(((((....)))))..))).))..((.(((((.....(((.....)))..))))))).......(((((.(.((((((((((....)))))))))).)))))) ( -41.70) >DroSec_CAF1 49352 96 + 1 ------------------GGGGGCAAACUUAAGAACAGCUGGCACUCGACACCGGACUUUAAGUCAAUCGAGGCAUAAAAAGUGUGCACAGGAGAAGCUCAAGAGUU------UCCACAU ------------------((..((..............(((((.(((((.....(((.....)))..)))))((((.....)))))).)))(((...)))....)).------.)).... ( -24.50) >DroSim_CAF1 57592 102 + 1 ------------------GGGGGCAAACUUAAGAACAGCUGGCACUCGACACUGGACUUUAAGUCAAUCGAGGCAUAAAAAGUGUGCACAGGAGAAGCUCAAGAGUUUUUCCUUCCACAU ------------------.((((((.((((.((.....)).((.(((((.....(((.....)))..))))))).....)))).)))..((((((((((....))))))))))))).... ( -29.90) >DroEre_CAF1 50679 104 + 1 GGUGGGGAU----------------GCCUUGAGAACAGACGGCACUCGACACUGGACUUUAAGUCAAUCGAGGCAUAAAAAGUGUGCGCAGGAGGAGCUCAAGAGUGUUUCCUUCCACAG ((.((..((----------------((((((((..(...(.((.(((((.....(((.....)))..)))))(((((.....))))))).)...)..)))))).))))..))..)).... ( -36.60) >DroYak_CAF1 53450 120 + 1 GAUGCGGAUAGGGCAGGUGGGUUAAGCCUUAUAAAAUGCCGGCACUCGACACUGGACUUUAAGUCAAUCGAGGCAUAAAAAGUGUGCGCAGGAGAAGCUCAAGAGUUUUUCCUUCCACAU .(((.(((...((((.(((((......)))))....)))).((.(((((.....(((.....)))..)))))(((((.....)))))))((((((((((....))))))))))))).))) ( -39.70) >consensus ___G_GG_U_________GGGGGCAACCUUAAGAACAGCUGGCACUCGACACUGGACUUUAAGUCAAUCGAGGCAUAAAAAGUGUGCGCAGGAGAAGCUCAAGAGUUUUUCCUUCCACAU .........................................((.(((((.....(((.....)))..))))))).......(((((...((((((((((....))))))))))..))))) (-22.86 = -24.34 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:41 2006