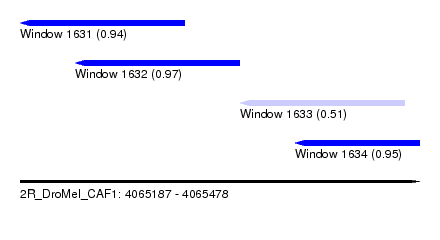
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,065,187 – 4,065,478 |
| Length | 291 |
| Max. P | 0.967051 |

| Location | 4,065,187 – 4,065,307 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -32.64 |
| Energy contribution | -33.44 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4065187 120 - 20766785 GGGGGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCGGCCACGCCCCCUGAAAGGCUGGCUGUUCAAACAUUUUUUUAUGUGUUUUAUACAAAUGCCUUGUCUUUGCGGGCCAUACUGAG ((((...(((.((((.......))))..))).))))((((.(((......((((((.((((((..((((((........))))))...)))...)))..)))))))))))))........ ( -38.90) >DroSec_CAF1 47789 116 - 1 GG-GGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCAGCCACGCCCCCUGAAAGGCUGGCUGUUCAAACAUU---UUAUGUGUUUUAUACAAAUGCCUUGUCUUUGCGGGCCAUACUGAG ((-((...)))).((.......))((((((....))))))..((((....((((((.((((((..((((((.---....))))))...)))...)))..))))))..))))......... ( -37.00) >DroSim_CAF1 54354 120 - 1 GGGGGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCAGCCACGCCCCCGGAAAGGCUGGCUGUUCAAACAUUUUUUUAUGUGUUUUAUACAAAUGCCUUGUCUUUGCGAGCCAUACUGAG (((((........((.......))((((((....))))))...)))))(.((((((.((((((..((((((........))))))...)))...)))..)))))).)............. ( -38.80) >DroEre_CAF1 49091 120 - 1 GGGGGAAACCCCUGCACAAUCUCCGGCUGAAACCUCAGCCACGCCCCCGGAAAGGCUGGCUGUUCAAACAUUUUUUUAUGUGUUUUAUACAAAUGCCUUGUCUUUGUGGGCCAUGCCGAG ((((....)))).(((........((((((....)))))).....((((.((((((.((((((..((((((........))))))...)))...)))..)))))).))))...))).... ( -45.90) >DroYak_CAF1 51911 120 - 1 GGGGGAAUCCCCCGCUAAAUCUCCGGGUGGAACCUCAGCCACGCCCCCGGAAAGGCUGCCUGUUCAAACAUUUUUUUAUGUGUUUUAUACAAAUGCCUUGUCUUUGUGGGCUUCACUGAG (((((...)))))..........(((((((........))))((((.(((((((((....(((..((((((........))))))...)))...)))))...)))).))))....))).. ( -37.30) >consensus GGGGGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCAGCCACGCCCCCGGAAAGGCUGGCUGUUCAAACAUUUUUUUAUGUGUUUUAUACAAAUGCCUUGUCUUUGCGGGCCAUACUGAG ((((....))))............((((((....)))))).....((((.((((((.((((((..((((((........))))))...)))...)))..)))))).)))).......... (-32.64 = -33.44 + 0.80)

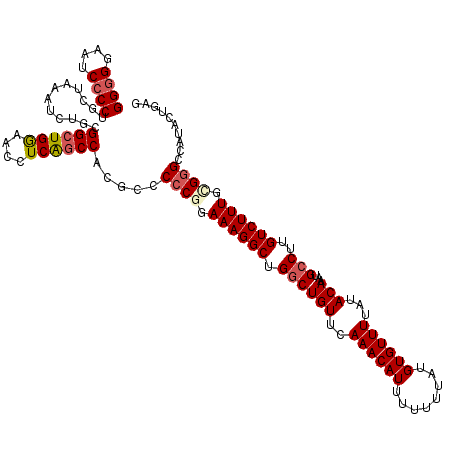
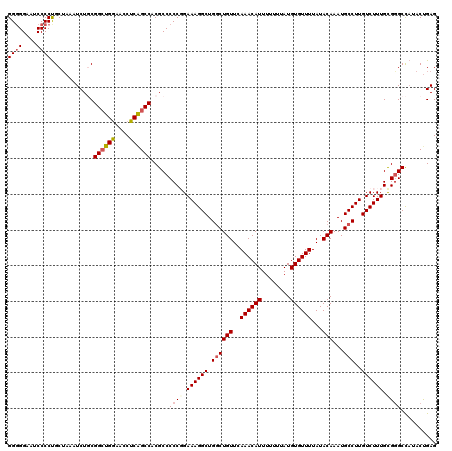
| Location | 4,065,227 – 4,065,347 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -34.28 |
| Energy contribution | -34.40 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4065227 120 - 20766785 AUUUAUAGUGUAUGACCGGCAAAUGUAGUUUGUCCCUGGCGGGGGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCGGCCACGCCCCCUGAAAGGCUGGCUGUUCAAACAUUUUUUUAUG ......(((((.((((((((....((((.((.....(((((((((...))))))))))).)))).))))).....((((((.(((........))))))))).))).)))))........ ( -42.20) >DroSec_CAF1 47829 116 - 1 AUUUAUAGUGUAUGACCGGCAAAUGUAGCUCGUCCCUGGCGG-GGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCAGCCACGCCCCCUGAAAGGCUGGCUGUUCAAACAUU---UUAUG ...((.(((((.((((((((....((((........((((((-((....))))))))...)))).))))).....((((((.(((........))))))))).))).)))))---.)).. ( -40.80) >DroSim_CAF1 54394 120 - 1 AUUUAUAGUGUAUGACCGGCAAAUGUAGCUCGUCCCUGGCGGGGGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCAGCCACGCCCCCGGAAAGGCUGGCUGUUCAAACAUUUUUUUAUG ......(((((.(((.((((.....(((((..(((.(((((((((...))))))))).....(.((((((....)))))).)......)))..))))))))).))).)))))........ ( -43.90) >DroEre_CAF1 49131 120 - 1 AUUUAUAGUGUAUGGCCGGCAAAUGUAGUUUGUCCCUGGCGGGGGAAACCCCUGCACAAUCUCCGGCUGAAACCUCAGCCACGCCCCCGGAAAGGCUGGCUGUUCAAACAUUUUUUUAUG ......(((((.((..((((.....((((((.(((..(((((((....))))............((((((....))))))..)))...))).))))))))))..)).)))))........ ( -44.30) >DroYak_CAF1 51951 120 - 1 AUUUAUAGUGUAUGGCUGGCAAAUGUAGUUUGUGCCUGGCGGGGGAAUCCCCCGCUAAAUCUCCGGGUGGAACCUCAGCCACGCCCCCGGAAAGGCUGCCUGUUCAAACAUUUUUUUAUG ......(((((.((((.((((............((((((((((((...)))))))).....((((((.((.............)))))))).)))))))).).))).)))))........ ( -42.42) >consensus AUUUAUAGUGUAUGACCGGCAAAUGUAGUUUGUCCCUGGCGGGGGAAUCCCCUGCUAAAUCUGCGGCUGGAACCUCAGCCACGCCCCCGGAAAGGCUGGCUGUUCAAACAUUUUUUUAUG ......(((((.(((.((((....((..........(((((((((...))))))))).......((((((....))))))))(((........)))..)))).))).)))))........ (-34.28 = -34.40 + 0.12)

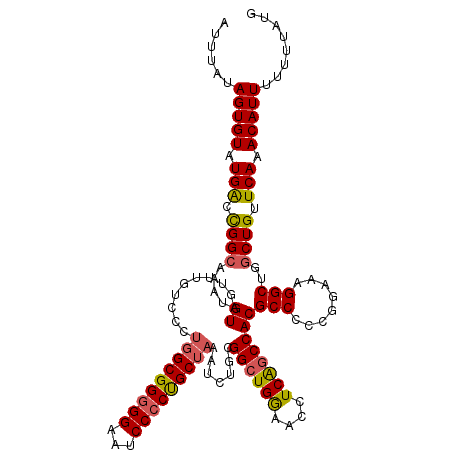
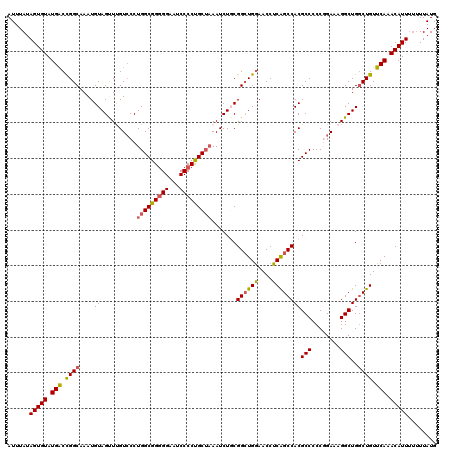
| Location | 4,065,347 – 4,065,467 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -38.66 |
| Consensus MFE | -30.86 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4065347 120 - 20766785 CGAGGAGCCAGGAUGUGCGGAUACGGAGUCCUGGGGAGUGGUGGGGAUUGCUUCAAUUCCGGGCUUGCCUUUUAGCGCAGCGCAAUAAAAAGGCAUAAAAAUGUUGAGCAUGUGGCGCUU ...(((((((((((...((....))..)))))))((((..((....))..))))..))))(.(((........))).)(((((.(((....(((((....))))).....))).))))). ( -39.10) >DroSec_CAF1 47945 117 - 1 CGAGGAGCCAGGAUGUGCGGAUACGAAGUCCUGGGGAGU---GGGAAUUGCUUCAAUUCCGGGCUUGCCUUUUAGCGCAACGUAAUAAAACGGCAUAAAAAUGUUGAGCAUGUGGCGCCU ..(((.((((.(.(((((((((.....)))).((.((((---.(((((((...))))))).).))).)).....)))))...........((((((....))))))....).)))).))) ( -39.10) >DroSim_CAF1 54514 117 - 1 CGAGGAGCCAGGAUGUGCGGAUACGAAGUCCUGGGGAGU---GGGAAUUGCUUCAAUUCCGGGCUUGCCUUUUAGCGCAACGUAAUAAAACGGCAUAAAAAUGUUGAGCAUGUGGCGCUU .((((..(((((((...((....))..))))))).((((---.(((((((...))))))).).))).))))..(((((.((((.......((((((....))))))...)))).))))). ( -38.80) >DroEre_CAF1 49251 117 - 1 CGAGGAUCCAGGAUGUGCGGAUGCGGAGUCCUGUGGACU---GGGAAUUGGUUCAAUUCCGGGCCUGCCUUUUAGCGCAAUGCAAUAAAACAGCAUAAAAAUGUUGAGCAUGUGGCGCUU .((((...((((((.(((....)))..)))))).((.((---.(((((((...))))))).))))..))))..(((((.((((.......((((((....)))))).))))...))))). ( -40.20) >DroYak_CAF1 52071 117 - 1 CGAGGAUCCAGGAUAUGCGGAUGCGGAGUCCUGUGGACU---CGGAAUUGGUUCAAUUUCGGGCUUGCCUUUUAGCGCAACGUAAUAAAACAGCAUAAAAAUGUUGAGCAUGUGGCGCUU .((((.((((((((.(((....)))..)))))).)).((---((((((((...))))))))))....))))..(((((.((((.......((((((....))))))...)))).))))). ( -36.10) >consensus CGAGGAGCCAGGAUGUGCGGAUACGGAGUCCUGGGGAGU___GGGAAUUGCUUCAAUUCCGGGCUUGCCUUUUAGCGCAACGUAAUAAAACGGCAUAAAAAUGUUGAGCAUGUGGCGCUU .((((..(((((((...((....))..))))))).........(((((((...))))))).......))))...((((.((((.......((((((....))))))...)))).)))).. (-30.86 = -30.98 + 0.12)

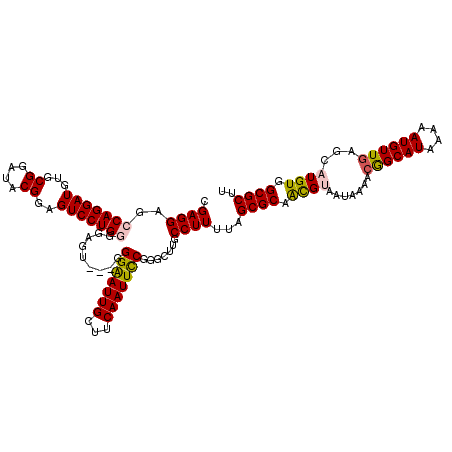
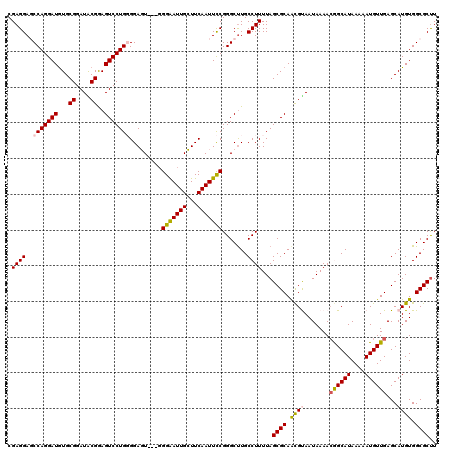
| Location | 4,065,387 – 4,065,478 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -26.17 |
| Energy contribution | -27.80 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4065387 91 - 20766785 CAGGGGAGAGGCGAGGAGCCAGGAUGUGCGGAUACGGAGUCCUGGGGAGUGGUGGGGAUUGCUUCAAUUCCGGGCUUGCCUUUUAGCGCAG ..(..((((((((((...(((((((...((....))..)))))))(((((..(((((....))))))))))...))))))))))..).... ( -35.80) >DroSec_CAF1 47985 88 - 1 CAGAGGAGAGGCGAGGAGCCAGGAUGUGCGGAUACGAAGUCCUGGGGAGU---GGGAAUUGCUUCAAUUCCGGGCUUGCCUUUUAGCGCAA ..(..((((((((((...(((((((...((....))..))))))).....---.(((((((...)))))))...))))))))))..).... ( -37.10) >DroEre_CAF1 49291 82 - 1 CA------AGGCGAGGAUCCAGGAUGUGCGGAUGCGGAGUCCUGUGGACU---GGGAAUUGGUUCAAUUCCGGGCCUGCCUUUUAGCGCAA .(------((((.(((.((((((((.(((....)))..)))))).)).((---.(((((((...))))))).))))))))))......... ( -29.90) >DroYak_CAF1 52111 83 - 1 CA-----GAGGCGAGGAUCCAGGAUAUGCGGAUGCGGAGUCCUGUGGACU---CGGAAUUGGUUCAAUUUCGGGCUUGCCUUUUAGCGCAA .(-----((((((((..((((((((.(((....)))..)))))).)).((---((((((((...)))))))))))))))))))........ ( -32.00) >consensus CA_____GAGGCGAGGAGCCAGGAUGUGCGGAUACGGAGUCCUGGGGACU___GGGAAUUGCUUCAAUUCCGGGCUUGCCUUUUAGCGCAA .....((((((((((...(((((((...((....))..))))))).........(((((((...)))))))...))))))))))....... (-26.17 = -27.80 + 1.63)

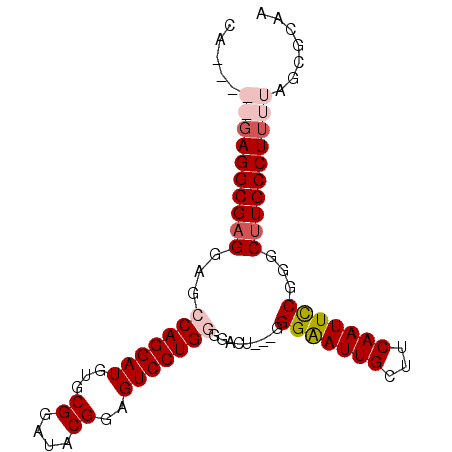
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:33 2006