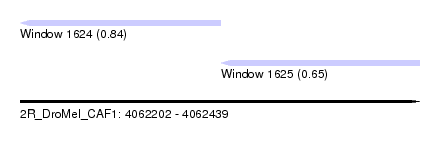
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,062,202 – 4,062,439 |
| Length | 237 |
| Max. P | 0.836888 |

| Location | 4,062,202 – 4,062,321 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -27.72 |
| Energy contribution | -27.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
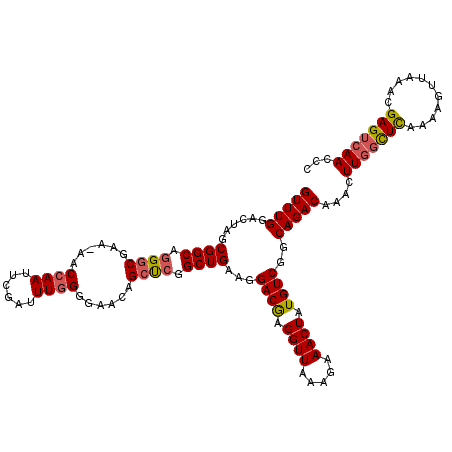
>2R_DroMel_CAF1 4062202 119 - 20766785 GUUUGGACUAGCGGCAGGACGAA-AACCAAUUCGAUUUGGGGAAAAGCUCGGCUGAAGGACGAGGUUAAAGAAACUAUGUCGGCAGACAAACUUGGCUCAAAAGUUAAACGAGUCAACCC (((((..((..((((.(..(...-..((((......))))......)..).)))).))((((.((((.....)))).))))..)))))....(((((((...........)))))))... ( -28.00) >DroSec_CAF1 44919 120 - 1 GUUUGGACUAGCGGCAGGGCCAAAAACCAAUUCGAUUUGGGGAACAGCCCGGCUGAAGGACGAGGUUAAAGAAACUAUGUCGGCAGACAAACUUGGCUCAAAAGUUAAACGAGGCAACCC (((((..((..((((.((((......((((......))))......)))).)))).))((((.((((.....)))).))))..)))))....(((.(((...........))).)))... ( -33.90) >DroSim_CAF1 51531 120 - 1 GUUUGGACGAGCGGCAGGGCCAAAACCCAAUUCGAUUUGGGGAACAGCUCGGCUGAAGGACGAGGUUAAAGAAACUAUGUCGGCAGACAAACUUGGCUCAAAAGUUAAACGAGUCAACCC (((((..((((((((...)))....(((((......))))).....))))).(((...((((.((((.....)))).))))..))).)))))(((((((...........)))))))... ( -39.20) >DroEre_CAF1 46083 119 - 1 GUUUGGACAAACGGCAGGGCGAA-AACCAAUUCGGUUUUGGGAACAGCUCGGCUGAAGGACAAGGUUAAGGAAACUAUGUCGGCAGACAAACUUGGCUUAAAAGUUAAACGAGUCAACCC (((((......((((.((((.((-((((.....))))))(....).)))).))))...((((.((((.....)))).))))..)))))....(((((((...........)))))))... ( -32.90) >DroYak_CAF1 48854 119 - 1 GUUUGGACUAACGGCAAGGCGAA-AACCAAUUCAGUUUGGGAAAAAGCUCGGCUGAAGGACAAGGUUAAAGAAACUAUGUCGGCAGACAAACUUCGUUUAAAAGUUAAACGAGUCAACCA (((((......((((..(((...-..((((......))))......)))..))))...((((.((((.....)))).))))..)))))....((((((((.....))))))))....... ( -31.70) >consensus GUUUGGACUAGCGGCAGGGCGAA_AACCAAUUCGAUUUGGGGAACAGCUCGGCUGAAGGACGAGGUUAAAGAAACUAUGUCGGCAGACAAACUUGGCUCAAAAGUUAAACGAGUCAACCC (((((......((((.((((......((((......))))......)))).))))...((((.((((.....)))).))))..)))))....(((((((...........)))))))... (-27.72 = -27.92 + 0.20)

| Location | 4,062,321 – 4,062,439 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.78 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
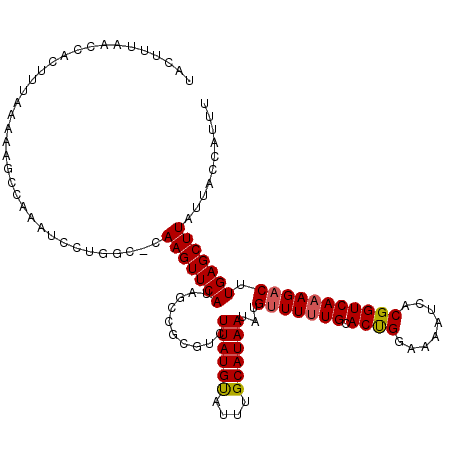
>2R_DroMel_CAF1 4062321 118 - 20766785 -GCUUUAACCACUUUAAAAAACCAAAUCCUGGC-CAAGUUCAUAGCCGCGUCUUAUGUAUUUGCAUAAUAUGUUUUUGCACUGGAAAAUCACGGUCAAAGACUUGAGCUUAUUACCAUUU -............................(((.-.(((((((..........((((((....))))))...(((((((.((((........))))))))))).)))))))....)))... ( -21.50) >DroSec_CAF1 45039 119 - 1 UACUUUAACCUCUUUAAAAAGCGAAAUCCUGGC-CAAGUUCAUAGCCGCGUCUUAUGUAUUUGCAUAAUAUGUUUUUGCACUGGAAAAUCACGGUCAAAGACUUGAGCUUAUUACCAUUU .............................(((.-.(((((((..........((((((....))))))...(((((((.((((........))))))))))).)))))))....)))... ( -21.50) >DroSim_CAF1 51651 120 - 1 UACUUUAACCACUUUAAAAAGCCAAAUCCUGGCACAAGUUCAUAGCCGCGUCUUAUGUAUUUGCAUAAUAUGCUUUUGCACUGGAAAAUCACGGUCAAAGACUUGAGCUUAUUACCAUUU ....................((((.....))))..(((((((.....((((.((((((....)))))).))))(((((.((((........)))))))))...))))))).......... ( -27.20) >DroEre_CAF1 46202 117 - 1 UACUUUAACCACUUUAA--AACCAAAUCCCAGC-AAAGUUCAUUGGCGAGUCUUAUGCAUUUGCAUAAUAUGUUUUUGCACUGGAAAAUCACGGUCAAAGACUUGAGCUUAUUACCAUUU .................--..............-.(((((((..........((((((....))))))...(((((((.((((........))))))))))).))))))).......... ( -22.30) >DroYak_CAF1 48973 117 - 1 UACUUUAAACACUUUAA--AGCCAAAUCCUGAC-AAAGUUCAUAGCCGAGUCUUAUGUAUUUGCAUAAUAUGUUUCUGCACCGAAAAAUCGCGGUCAAAGACUUGAGCUUAUUACCAUUU ..((((((.....))))--))............-...((....(((((((((((((((....))))..........((.((((........)))))))))))))).)))....))..... ( -21.60) >consensus UACUUUAACCACUUUAAAAAGCCAAAUCCUGGC_CAAGUUCAUAGCCGCGUCUUAUGUAUUUGCAUAAUAUGUUUUUGCACUGGAAAAUCACGGUCAAAGACUUGAGCUUAUUACCAUUU ...................................(((((((..........((((((....))))))...(((((((.((((........))))))))))).))))))).......... (-19.62 = -19.70 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:24 2006