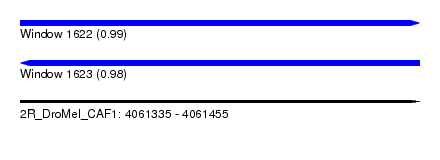
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,061,335 – 4,061,455 |
| Length | 120 |
| Max. P | 0.990483 |

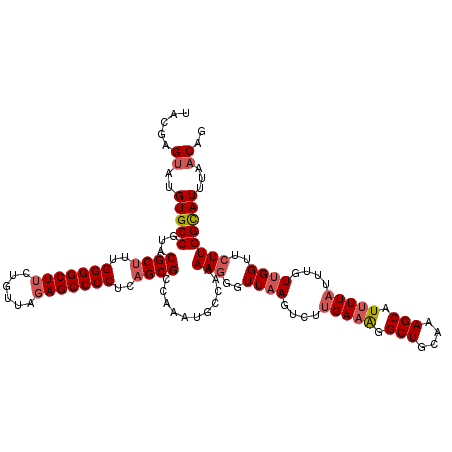
| Location | 4,061,335 – 4,061,455 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -27.46 |
| Energy contribution | -28.50 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4061335 120 + 20766785 CUGUUAAAUGCCAAGAACCAACAAAGAAAAUGCUUUGCAGCCUUUCAAGACUUAACCCUUUUGACAUUUGGCGCUGAGGGGCUCUAACAGAAGCCCCAAGGCGUACGGCACAUACUCGUA ........((((.........(((((......)))))..(((..((((((........)))))).....)))(((..(((((((.....).))))))..)))....)))).......... ( -32.30) >DroSec_CAF1 44040 119 + 1 CUGUUAAAUGCCAAGAACCAACAAAUAAAAUGCUUUGCAGCCUUUCAAGACUUAACCCUUUUGGCAUUU-GCGCUGAGGGGCUCUAACAGAAGCCCCAAAGCGUGCGGCACAUACUCGUA ((((.(((((((((((...........(((.(((....))).))).............)))))))))))-(((((..(((((((.....).))))))..)))))))))............ ( -33.35) >DroSim_CAF1 50652 120 + 1 CUGUUAAAUGCCAACAACCAACAAAUAAAAUGCUUUGCAGCCUUUCAAGACUUAACCCUUUUGGCAUUUGGCGCUGAGGGGCUCUAAUAGAAGCCCCAAAGCGUGCGGCACAUACUCGUA ........((((.........(((((....(((...)))(((....(((........)))..))))))))(((((..(((((((.....).))))))..)))))..)))).......... ( -34.20) >DroEre_CAF1 45232 119 + 1 CUGUUAAAUGCCAAGAACCAACAAAUAA-AAGCUUUGCAGCCCUUCAAGACUAAACCCUUUUGGCAUUGGGCGCUUUGGGGCUAUAAGAGAAGCCCCAAAGCGUAGGGCACAUGCUUGUA ............................-((((..((..(((((((((..(((((....)))))..))))(((((((((((((........)))))))))))))))))).)).))))... ( -41.30) >DroYak_CAF1 47984 120 + 1 CUGUUAAAUACCAAGAACCAACAAAUAAAAAGCUUUGAAGCCUUUCACGACUUAACCCUUUUGCCAUUUCGCGCUAAGGGGCUCCAACAGAAGCCCCAAAGCGUACGGCACAUACUCGUA ..............((...............(((....)))....................((((.....(((((..((((((.(....).))))))..)))))..)))).....))... ( -24.50) >consensus CUGUUAAAUGCCAAGAACCAACAAAUAAAAUGCUUUGCAGCCUUUCAAGACUUAACCCUUUUGGCAUUUGGCGCUGAGGGGCUCUAACAGAAGCCCCAAAGCGUACGGCACAUACUCGUA ..((((((((((((((...............(((....)))(......).........))))))))))))))(((..((((((........))))))..))).(((((.......))))) (-27.46 = -28.50 + 1.04)

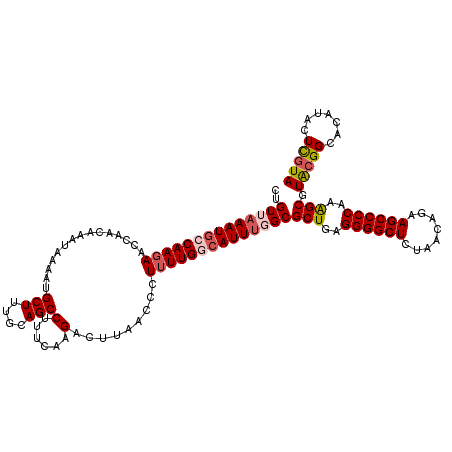
| Location | 4,061,335 – 4,061,455 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.84 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4061335 120 - 20766785 UACGAGUAUGUGCCGUACGCCUUGGGGCUUCUGUUAGAGCCCCUCAGCGCCAAAUGUCAAAAGGGUUAAGUCUUGAAAGGCUGCAAAGCAUUUUCUUUGUUGGUUCUUGGCAUUUAACAG ((((.((....))))))(((...(((((((......)))))))...)))..((((((((..((..((((((((....)))))((((((......)))))))))..))))))))))..... ( -36.90) >DroSec_CAF1 44040 119 - 1 UACGAGUAUGUGCCGCACGCUUUGGGGCUUCUGUUAGAGCCCCUCAGCGC-AAAUGCCAAAAGGGUUAAGUCUUGAAAGGCUGCAAAGCAUUUUAUUUGUUGGUUCUUGGCAUUUAACAG .....((..((((((((((((..(((((((......)))))))..)))).-...)))...(((..((((....(((((.(((....))).)))))....))))..))))))))...)).. ( -39.40) >DroSim_CAF1 50652 120 - 1 UACGAGUAUGUGCCGCACGCUUUGGGGCUUCUAUUAGAGCCCCUCAGCGCCAAAUGCCAAAAGGGUUAAGUCUUGAAAGGCUGCAAAGCAUUUUAUUUGUUGGUUGUUGGCAUUUAACAG .....((..((((((.(((((..(((((((......)))))))..)))(((((..(((.....))).......(((((.(((....))).)))))....))))).))))))))...)).. ( -37.90) >DroEre_CAF1 45232 119 - 1 UACAAGCAUGUGCCCUACGCUUUGGGGCUUCUCUUAUAGCCCCAAAGCGCCCAAUGCCAAAAGGGUUUAGUCUUGAAGGGCUGCAAAGCUU-UUAUUUGUUGGUUCUUGGCAUUUAACAG ...((((.((((((((.((((((((((((........)))))))))))((((..........))))........).))))).)))..))))-(((..(((..(...)..)))..)))... ( -42.50) >DroYak_CAF1 47984 120 - 1 UACGAGUAUGUGCCGUACGCUUUGGGGCUUCUGUUGGAGCCCCUUAGCGCGAAAUGGCAAAAGGGUUAAGUCGUGAAAGGCUUCAAAGCUUUUUAUUUGUUGGUUCUUGGUAUUUAACAG ...((((((.((((((.((((..((((((((....))))))))..))))....)))))).(((..((((...((((((((((....))))))))))...))))..))).))))))..... ( -46.80) >consensus UACGAGUAUGUGCCGUACGCUUUGGGGCUUCUGUUAGAGCCCCUCAGCGCCAAAUGCCAAAAGGGUUAAGUCUUGAAAGGCUGCAAAGCAUUUUAUUUGUUGGUUCUUGGCAUUUAACAG .....((..(((((...((((..(((((((......)))))))..))))...........(((..((((....(((((.(((....))).)))))....))))..))))))))...)).. (-31.76 = -32.84 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:22 2006