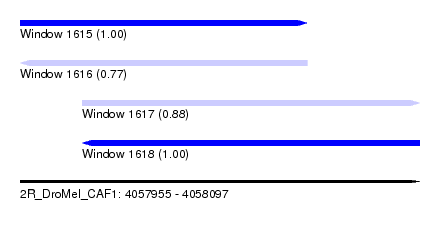
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,057,955 – 4,058,097 |
| Length | 142 |
| Max. P | 0.999428 |

| Location | 4,057,955 – 4,058,057 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4057955 102 + 20766785 ------------CAAAGCCAAAGCCAUAG------CAAACCGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAUGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUU ------------.........((((((.(------(...(((((.(((...(..((((((((....(((......))).))))))))..).)))..)))))(((....))))).)))))) ( -31.60) >DroSec_CAF1 40651 106 + 1 --------AAGCCAAAGCCAAAGCCAUAG------CAAACCGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUU --------.............((((((.(------(...(((((.(((...(..((((((((.....((......))..))))))))..).)))..)))))(((....))))).)))))) ( -30.90) >DroSim_CAF1 46837 106 + 1 --------AAGCCAAAGCCAAAGCCAUAG------CAAACCGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUU --------.............((((((.(------(...(((((.(((...(..((((((((.....((......))..))))))))..).)))..)))))(((....))))).)))))) ( -30.90) >DroEre_CAF1 41474 116 + 1 GAAAGCCAAAGCCAAAGCCAAAGCCAAAGCAAGACCAAACCGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGG----GCUCGCAAUGGUU ...(((((..((...((((...((....)).........(((((.(((...(..((((((((.....((......))..))))))))..).)))..))))).)----))).))..))))) ( -31.70) >DroYak_CAF1 44193 112 + 1 --------AAGCCAAAGCCAAAGCCAAAGCAAGACCAAACCGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUU --------.............(((((..((.........(((((.(((...(..((((((((.....((......))..))))))))..).)))..)))))(((....)))))..))))) ( -28.90) >consensus ________AAGCCAAAGCCAAAGCCAUAG______CAAACCGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUU ...............(((((..((...............(((((.(((...(..((((((((.(((......)))....))))))))..).)))..)))))(((....)))))..))))) (-26.68 = -26.72 + 0.04)

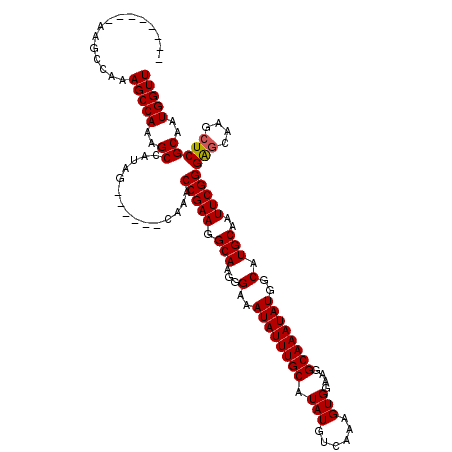
| Location | 4,057,955 – 4,058,057 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -20.05 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4057955 102 - 20766785 AACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCAUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCGGUUUG------CUAUGGCUUUGGCUUUG------------ ...((..(((((((.((..(((((..(((....((((((((.................))))))))......))).)))))...)------)...)))))..))..))------------ ( -22.93) >DroSec_CAF1 40651 106 - 1 AACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCGGUUUG------CUAUGGCUUUGGCUUUGGCUU-------- ..(((..(((((((.((..(((((..(((....((((((((.................))))))))......))).)))))...)------)...)))))..))..)))...-------- ( -26.83) >DroSim_CAF1 46837 106 - 1 AACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCGGUUUG------CUAUGGCUUUGGCUUUGGCUU-------- ..(((..(((((((.((..(((((..(((....((((((((.................))))))))......))).)))))...)------)...)))))..))..)))...-------- ( -26.83) >DroEre_CAF1 41474 116 - 1 AACCAUUGCGAGC----CCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCGGUUUGGUCUUGCUUUGGCUUUGGCUUUGGCUUUGGCUUUC ...((..((((((----(.(((((..(((....((((((((.................))))))))......))).)))))...)).)))))..))(((..(((....)))..))).... ( -31.13) >DroYak_CAF1 44193 112 - 1 AACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCGGUUUGGUCUUGCUUUGGCUUUGGCUUUGGCUU-------- ..(((..(((((((.....(((((..(((....((((((((.................))))))))......))).)))))...)).)))))..)))....(((....))).-------- ( -26.13) >consensus AACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCGGUUUG______CUAUGGCUUUGGCUUUGGCUU________ ..(((..(((((....)))(((((..(((....((((((((.................))))))))......))).)))))...............))..)))................. (-20.05 = -20.45 + 0.40)

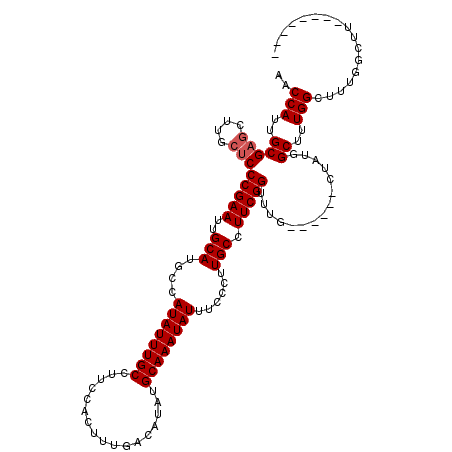
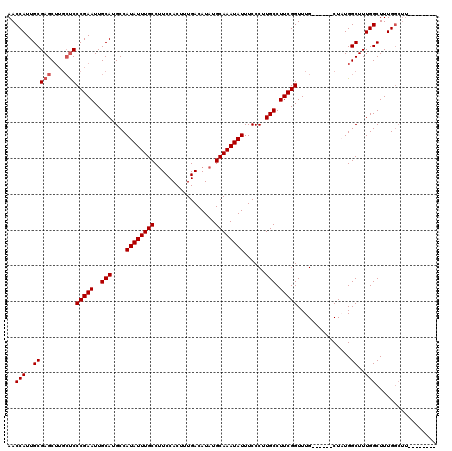
| Location | 4,057,977 – 4,058,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -32.50 |
| Energy contribution | -32.74 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4057977 120 + 20766785 CGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAUGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUUGACUGCAUAAACCGCACUGCAUGGCCACUAACAAGUGCCG ....((((...(..((((((((....(((......))).))))))))((((((((...((((((....)))(((.........))).....)))...))))).))).....)...)))). ( -35.60) >DroSec_CAF1 40677 120 + 1 CGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUUGACUGCAUAAACCUCACUGCAUGGCCACUAACAAGUGCCG ....((((...(..((((((((.....((......))..))))))))((((((((....(.(((....))).)...((((.........))))....))))).))).....)...)))). ( -33.50) >DroSim_CAF1 46863 120 + 1 CGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUUGACUGCAUAAACCGCACUGCAUGGCCACUAACAAGUGCCG ....((((...(..((((((((.....((......))..))))))))((((((((...((((((....)))(((.........))).....)))...))))).))).....)...)))). ( -34.90) >DroEre_CAF1 41514 116 + 1 CGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGG----GCUCGCAAUGGUUGACUGCAUAAACCGCACUGCAUGGCCACUAACCAGUGCCG ....((((..((..((((((((.....((......))..))))))))((((((((...(((..----((...((.....))...)).....)))...))))).))).....))..)))). ( -35.60) >DroYak_CAF1 44225 120 + 1 CGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUUGACUGCAUAAACCGCACUGCAUGGCCACUAACAAGUGCCG ....((((...(..((((((((.....((......))..))))))))((((((((...((((((....)))(((.........))).....)))...))))).))).....)...)))). ( -34.90) >consensus CGAAGGCAAGGGAAAUAUUUGCAUAUGUCAAAGUGGAAGGCAAAUAUGGCAUGCAAUUCGGGAGCAAGCUCGCAAUGGUUGACUGCAUAAACCGCACUGCAUGGCCACUAACAAGUGCCG ....((((...(..((((((((.(((......)))....))))))))((((((((...((((((....)))(((.........))).....)))...))))).))).....)...)))). (-32.50 = -32.74 + 0.24)

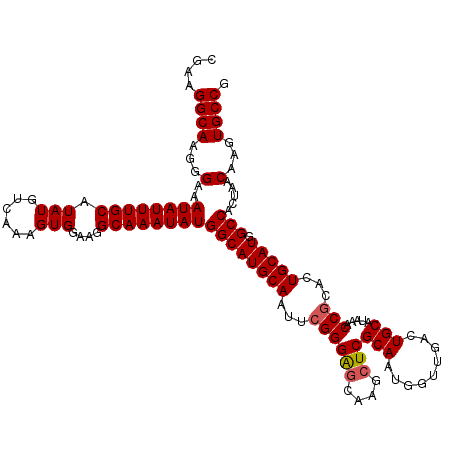
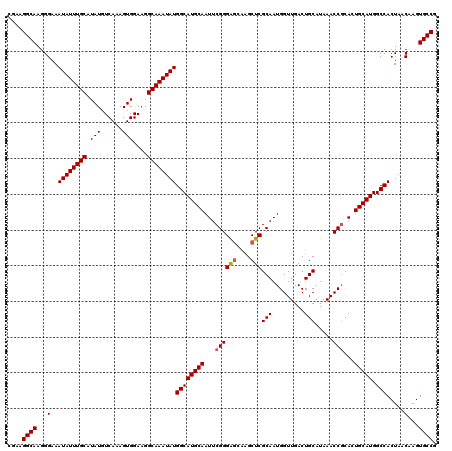
| Location | 4,057,977 – 4,058,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -32.55 |
| Energy contribution | -33.15 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4057977 120 - 20766785 CGGCACUUGUUAGUGGCCAUGCAGUGCGGUUUAUGCAGUCAACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCAUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCG .((((.......(((((.(((((((.(((.....(((((.....)))))(((....)))))).))))))))))))((((((.................))))))........)))).... ( -35.13) >DroSec_CAF1 40677 120 - 1 CGGCACUUGUUAGUGGCCAUGCAGUGAGGUUUAUGCAGUCAACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCG .((((.......(((((.(((((((..((.....(((((.....)))))(((....)))))..))))))))))))((((((.................))))))........)))).... ( -33.83) >DroSim_CAF1 46863 120 - 1 CGGCACUUGUUAGUGGCCAUGCAGUGCGGUUUAUGCAGUCAACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCG .((((.......(((((.(((((((.(((.....(((((.....)))))(((....)))))).))))))))))))((((((.................))))))........)))).... ( -35.13) >DroEre_CAF1 41514 116 - 1 CGGCACUGGUUAGUGGCCAUGCAGUGCGGUUUAUGCAGUCAACCAUUGCGAGC----CCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCG .((((..((...(((((.(((((((.(((....((((((.....))))))...----..))).))))))))))))((((((.................))))))....))..)))).... ( -34.53) >DroYak_CAF1 44225 120 - 1 CGGCACUUGUUAGUGGCCAUGCAGUGCGGUUUAUGCAGUCAACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCG .((((.......(((((.(((((((.(((.....(((((.....)))))(((....)))))).))))))))))))((((((.................))))))........)))).... ( -35.13) >consensus CGGCACUUGUUAGUGGCCAUGCAGUGCGGUUUAUGCAGUCAACCAUUGCGAGCUUGCUCCCGAAUUGCAUGCCAUAUUUGCCUUCCACUUUGACAUAUGCAAAUAUUUCCCUUGCCUUCG .((((.......(((((.(((((((.(((.....(((((.....)))))(((....)))))).))))))))))))((((((.................))))))........)))).... (-32.55 = -33.15 + 0.60)

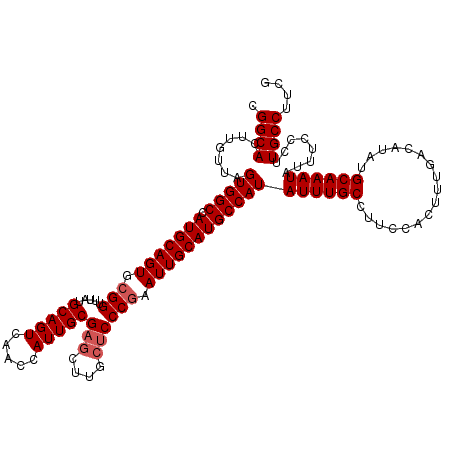
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:17 2006