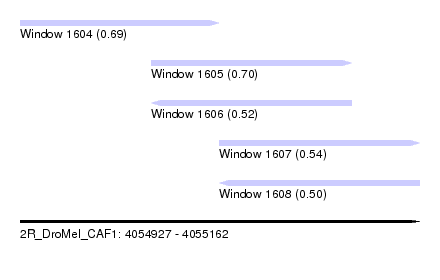
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,054,927 – 4,055,162 |
| Length | 235 |
| Max. P | 0.701166 |

| Location | 4,054,927 – 4,055,044 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4054927 117 + 20766785 CUUUUUUCCGC---UGCUGGCAAGUUGCCAUUGUCCGAUUACAUUCUGCCAAGCUGGCAUUCCAUUCCGGCACCAGCUGCUCCUCAUUCCCGCCCCCAUCCAUACACGUACCUCCACCAU .........((---.(((((.....((((.......((......))((((.....)))).........))))))))).))........................................ ( -21.90) >DroSim_CAF1 43713 117 + 1 CUUUUUUACGCUGCUGCUGGCAAGUUGCCAUUGUCCGAUUACAUUUUGCCAAGCUGGCAUUCCAUUCCGGCACCAGCUGCUCCUCAUUCCCGCCCCUAUCCAUACACACACCUCCAC--- .........((.((((.(((((((.((..(((....)))..)).))))))).(((((.(......))))))..)))).)).....................................--- ( -23.90) >DroEre_CAF1 37570 116 + 1 CUU-UUUCCGC---UGCUGGCAAGUUGCCAUUGUCCGACUACAUUCUGCCAAGCUGGCAUUCCAUUCCGGCACCAGCUGCUCCUCAUUCCCGGCCACAUCCAGGCACACACCUCCACCGC ...-.....((---.(((((.....((((..(((......)))...((((.....)))).........))))))))).))............(((.......)))............... ( -25.50) >DroYak_CAF1 41037 113 + 1 CUUUUUUCCGC---UGCUGGCAAGUUGCCAUUGUCCGAUUACAUUCUGCCAAGCUGGCAUUCCAUUCCGGCACCAGCUGCUCCUCAUACCCGUCCAUAUCAACAC----ACAUACACCUC .........((---.(((((.....((((.......((......))((((.....)))).........))))))))).)).........................----........... ( -21.90) >consensus CUUUUUUCCGC___UGCUGGCAAGUUGCCAUUGUCCGAUUACAUUCUGCCAAGCUGGCAUUCCAUUCCGGCACCAGCUGCUCCUCAUUCCCGCCCACAUCCAUACACACACCUCCACC_C .........((....(((((.....((((..(((......)))...((((.....)))).........))))))))).))........................................ (-19.50 = -19.50 + 0.00)

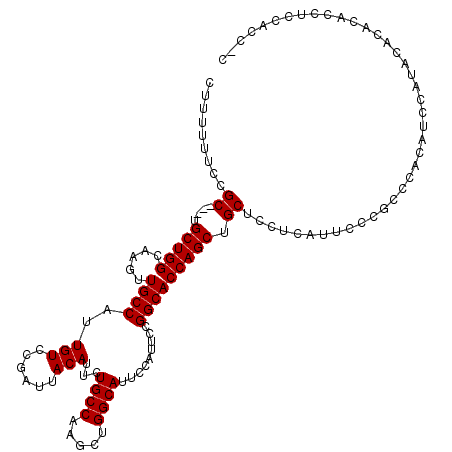
| Location | 4,055,004 – 4,055,122 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -11.20 |
| Energy contribution | -12.45 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4055004 118 + 20766785 UCCUCAUUCCCGCCCCCAUCCAUACACGUACCUCCACCAUCGCCACCAUCC--UCAGCCGUAUCCAUGCCCAUAUCCAGGCGAGUAUGCCCCACCCAAAAGGGCAGCAUAAAAACGGGGC ...........(((((.......................(((((.......--...((.........)).........))))).((((((((........)))..))))).....))))) ( -24.41) >DroSim_CAF1 43793 105 + 1 UCCUCAUUCCCGCCCCUAUCCAUACACACACCUCCAC------------CC--UCAGCCGUAUCCAUGCCCAUAUCCACGCCAGUAUGCC-CACCCAAAAGGGCAGCAUAAAAACGGGGC ...........(((((.....((((............------------..--......))))..((((........((....)).((((-(........)))))))))......))))) ( -20.65) >DroEre_CAF1 37646 117 + 1 UCCUCAUUCCCGGCCACAUCCAGGCACACACCUCCACCGCCACCACCACCCCAUAGCCCGUAUCUAUUCUCAUAUCCAGGCCAGUA--CC-CGCCCAAAAGGGCAGCAUAAAAACGGGGC .......((((((((.......(((.............)))...........((((.......))))...........))))....--..-.((((....))))...........)))). ( -25.22) >DroYak_CAF1 41114 107 + 1 UCCUCAUACCCGUCCAUAUCAACAC----ACAUACACCUCCACCC--------UUACCCGUAUCUAUGGCCAUAUCCAGGCCAGUAUGCC-CACCCAAAAGGGCAGCAUAAAAACGGGGC .........................----................--------...(((((.....(((((.......)))))((.((((-(........)))))))......))))).. ( -26.40) >consensus UCCUCAUUCCCGCCCACAUCCAUACACACACCUCCACC_CCACCA____CC__UCACCCGUAUCCAUGCCCAUAUCCAGGCCAGUAUGCC_CACCCAAAAGGGCAGCAUAAAAACGGGGC ........((((............................................(((((((........))))...)))...(((((....(((....)))..)))))....)))).. (-11.20 = -12.45 + 1.25)

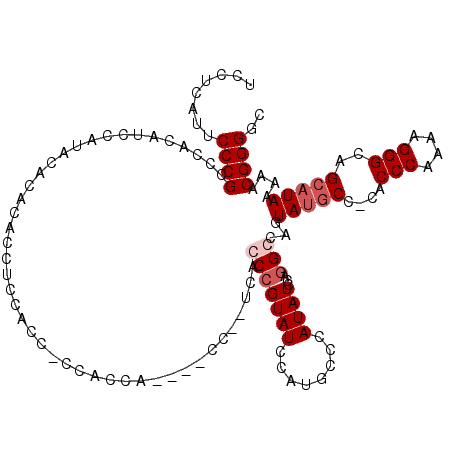
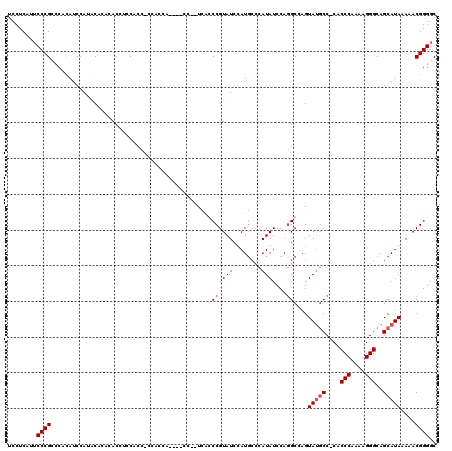
| Location | 4,055,004 – 4,055,122 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.75 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4055004 118 - 20766785 GCCCCGUUUUUAUGCUGCCCUUUUGGGUGGGGCAUACUCGCCUGGAUAUGGGCAUGGAUACGGCUGA--GGAUGGUGGCGAUGGUGGAGGUACGUGUAUGGAUGGGGGCGGGAAUGAGGA ..(((((((((((.((((((.((..((((((.....))))))..))...)))))...((((((((..--..((.((....)).))...))).)))))..).)))))))))))........ ( -42.50) >DroSim_CAF1 43793 105 - 1 GCCCCGUUUUUAUGCUGCCCUUUUGGGUG-GGCAUACUGGCGUGGAUAUGGGCAUGGAUACGGCUGA--GG------------GUGGAGGUGUGUGUAUGGAUAGGGGCGGGAAUGAGGA ((((((((..(((((..(((....)))..-).))))..)))...........((((.(((((.((..--..------------....)).))))).))))....)))))........... ( -37.50) >DroEre_CAF1 37646 117 - 1 GCCCCGUUUUUAUGCUGCCCUUUUGGGCG-GG--UACUGGCCUGGAUAUGAGAAUAGAUACGGGCUAUGGGGUGGUGGUGGCGGUGGAGGUGUGUGCCUGGAUGUGGCCGGGAAUGAGGA (((((((.......((((((....)))))-).--....((((((..(((....)))....)))))))))))))..((((.(((....((((....))))...))).)))).......... ( -43.40) >DroYak_CAF1 41114 107 - 1 GCCCCGUUUUUAUGCUGCCCUUUUGGGUG-GGCAUACUGGCCUGGAUAUGGCCAUAGAUACGGGUAA--------GGGUGGAGGUGUAUGU----GUGUUGAUAUGGACGGGUAUGAGGA ((((((..(((((.((((((........)-))))...(((((.......))))).......).))))--------)..))).)))((((.(----((.(......).))).))))..... ( -34.80) >consensus GCCCCGUUUUUAUGCUGCCCUUUUGGGUG_GGCAUACUGGCCUGGAUAUGGGCAUAGAUACGGCUGA__GG____UGGUGG_GGUGGAGGUGUGUGUAUGGAUAGGGGCGGGAAUGAGGA ..((((((((((((((((((....))))..)))....(((((((..(((....)))....)))))))..................................)))))))))))........ (-22.62 = -23.75 + 1.12)

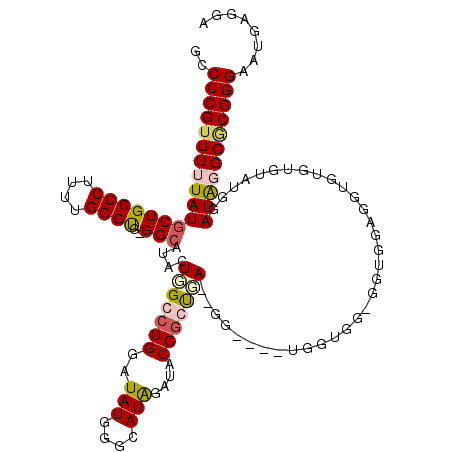
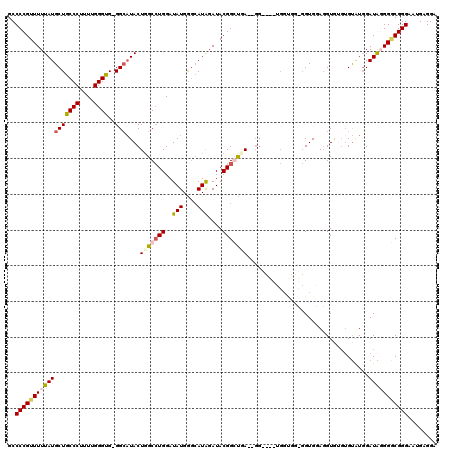
| Location | 4,055,044 – 4,055,162 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -22.15 |
| Energy contribution | -23.65 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4055044 118 + 20766785 CGCCACCAUCC--UCAGCCGUAUCCAUGCCCAUAUCCAGGCGAGUAUGCCCCACCCAAAAGGGCAGCAUAAAAACGGGGCAUCAUAAAAAAAUGGCGUCAAUUGGCGACGCGUUAGCCUG (((((......--...(((((.....((((........)))).(.(((((((.(((....)))..(........)))))))))........)))))......)))))..((....))... ( -34.06) >DroSim_CAF1 43830 108 + 1 ---------CC--UCAGCCGUAUCCAUGCCCAUAUCCACGCCAGUAUGCC-CACCCAAAAGGGCAGCAUAAAAACGGGGCAUCAUAAAAAAAUGGCGUCAAUUGGCGACGCGUUAGCCUG ---------..--....((((....((((........((....)).((((-(........)))))))))....))))(((..(((......)))(((((.......)))))....))).. ( -29.20) >DroEre_CAF1 37686 117 + 1 CACCACCACCCCAUAGCCCGUAUCUAUUCUCAUAUCCAGGCCAGUA--CC-CGCCCAAAAGGGCAGCAUAAAAACGGGGCAUCAUAAAAAAAUGGCGUCAAUUGGCGACGCGUUAGCCUG ....................................(((((..((.--((-(((((....)))).(........))))))..............(((((.......)))))....))))) ( -26.70) >DroYak_CAF1 41150 111 + 1 CACCC--------UUACCCGUAUCUAUGGCCAUAUCCAGGCCAGUAUGCC-CACCCAAAAGGGCAGCAUAAAAACGGGGCAUCAUAAAAAAAUGGCGUCAAUUGGCGACGCGUUAGCCUG .....--------....((((.....(((((.......)))))((.((((-(........)))))))......))))(((..(((......)))(((((.......)))))....))).. ( -35.50) >consensus CACCA____CC__UCACCCGUAUCCAUGCCCAUAUCCAGGCCAGUAUGCC_CACCCAAAAGGGCAGCAUAAAAACGGGGCAUCAUAAAAAAAUGGCGUCAAUUGGCGACGCGUUAGCCUG .................((((.(.(((((.........(((......)))...(((....)))..))))).).))))(((..(((......)))(((((.......)))))....))).. (-22.15 = -23.65 + 1.50)

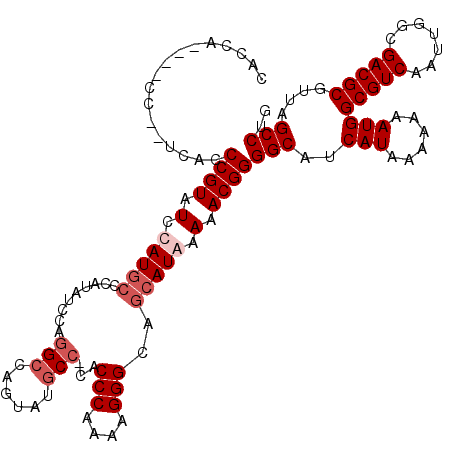
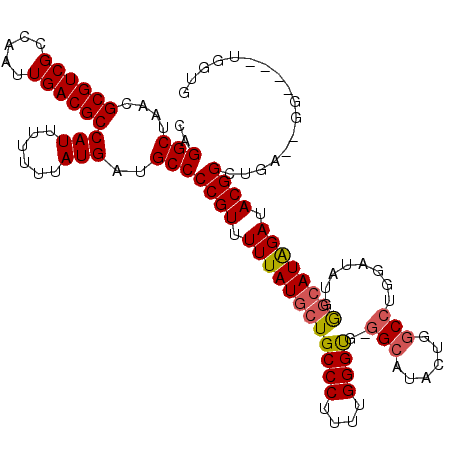
| Location | 4,055,044 – 4,055,162 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -31.85 |
| Energy contribution | -32.23 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4055044 118 - 20766785 CAGGCUAACGCGUCGCCAAUUGACGCCAUUUUUUUAUGAUGCCCCGUUUUUAUGCUGCCCUUUUGGGUGGGGCAUACUCGCCUGGAUAUGGGCAUGGAUACGGCUGA--GGAUGGUGGCG .........((((((.....))))))..............((((((((((((.(((((((.((..((((((.....))))))..))...))))..(....)))))))--)))))).))). ( -46.50) >DroSim_CAF1 43830 108 - 1 CAGGCUAACGCGUCGCCAAUUGACGCCAUUUUUUUAUGAUGCCCCGUUUUUAUGCUGCCCUUUUGGGUG-GGCAUACUGGCGUGGAUAUGGGCAUGGAUACGGCUGA--GG--------- (((.(....((((((.....))))))..........(.((((((((((..(((((..(((....)))..-).))))..)))).......)))))).)....).))).--..--------- ( -36.71) >DroEre_CAF1 37686 117 - 1 CAGGCUAACGCGUCGCCAAUUGACGCCAUUUUUUUAUGAUGCCCCGUUUUUAUGCUGCCCUUUUGGGCG-GG--UACUGGCCUGGAUAUGAGAAUAGAUACGGGCUAUGGGGUGGUGGUG ...((((.(((((((.....))))))..............(((((((.......((((((....)))))-).--....((((((..(((....)))....)))))))))))))).)))). ( -41.30) >DroYak_CAF1 41150 111 - 1 CAGGCUAACGCGUCGCCAAUUGACGCCAUUUUUUUAUGAUGCCCCGUUUUUAUGCUGCCCUUUUGGGUG-GGCAUACUGGCCUGGAUAUGGCCAUAGAUACGGGUAA--------GGGUG ..(((....((((((.....))))))(((......)))..)))((((.(((((.(..(((....)))..-).......((((.......))))))))).))))....--------..... ( -36.70) >consensus CAGGCUAACGCGUCGCCAAUUGACGCCAUUUUUUUAUGAUGCCCCGUUUUUAUGCUGCCCUUUUGGGUG_GGCAUACUGGCCUGGAUAUGGGCAUAGAUACGGCUGA__GG____UGGUG ..(((....((((((.....))))))(((......)))..)))((((.((((((((((((....))))..(((......)))........)))))))).))))................. (-31.85 = -32.23 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:07 2006