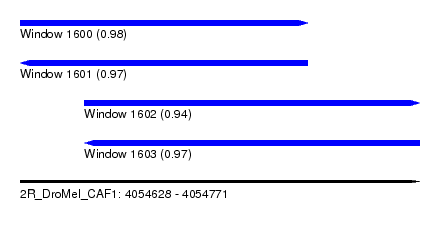
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,054,628 – 4,054,771 |
| Length | 143 |
| Max. P | 0.982764 |

| Location | 4,054,628 – 4,054,731 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.13 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4054628 103 + 20766785 AA-----------------AAAAUAGGAUAAGAUAGCGAUGUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCGUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUA ..-----------------((((((.(....((((((((.(((((.(((....)))))))))))).))))..).))))))....(((..(((((.(((....))).)))))..))).... ( -27.20) >DroSec_CAF1 37308 103 + 1 AA-----------------AAAAUAGCAUAAGAUAGCGAUGUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUA ..-----------------((((((((....((((((((.(((((.(((....)))))))))))).)))).)).))))))....(((..(((((.(((....))).)))))..))).... ( -27.60) >DroSim_CAF1 43434 103 + 1 AA-----------------AAAAUAGGAUAAGAUAGCGAUGUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUA ..-----------------((((((.(....((((((((.(((((.(((....)))))))))))).))))..).))))))....(((..(((((.(((....))).)))))..))).... ( -25.10) >DroEre_CAF1 37251 120 + 1 AAAAAUAGGGAAAAUAGGGAAAAUAGGAUAAGAUAGCGCUGUAUAAUAUGAAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCUGACAUGCCGGCAAAAUUA ................(..((((((.(....(((((((..(((((.(((....)))))))).))).))))..).))))))..).(((..(((((.(((....))).)))))..))).... ( -25.60) >consensus AA_________________AAAAUAGGAUAAGAUAGCGAUGUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUA ...................................(((((((..((((((..(((((........)))))..))))))...)))))...(((((.(((....))).)))))))....... (-22.86 = -22.93 + 0.06)


| Location | 4,054,628 – 4,054,731 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.13 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -21.51 |
| Energy contribution | -21.32 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4054628 103 - 20766785 UAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACACGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUACAUCGCUAUCUUAUCCUAUUUU-----------------UU .......(((((((((((....)))))))))...................((((((.(((.......))))))))).......))................-----------------.. ( -22.10) >DroSec_CAF1 37308 103 - 1 UAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUACAUCGCUAUCUUAUGCUAUUUU-----------------UU .......(((((((((((....)))))))))....((((....)))))).((((((.(((.......))))))))).......((........))......-----------------.. ( -23.20) >DroSim_CAF1 43434 103 - 1 UAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUACAUCGCUAUCUUAUCCUAUUUU-----------------UU .......((.(((((((.....(((((........)))))...)))))))((((((.(((.......))))))))).......))................-----------------.. ( -22.60) >DroEre_CAF1 37251 120 - 1 UAAUUUUGCCGGCAUGUCAGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUUCAUAUUAUACAGCGCUAUCUUAUCCUAUUUUCCCUAUUUUCCCUAUUUUU .......((.(((((((.....(((((........)))))...)))))))((((((.((((....)))).)))))).....))..................................... ( -22.10) >consensus UAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUACAUCGCUAUCUUAUCCUAUUUU_________________UU .......(((((((((((....)))))))))...................((((((.(((.......))))))))).......))................................... (-21.51 = -21.32 + -0.19)

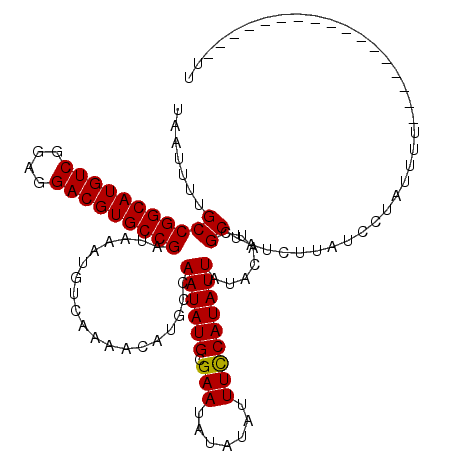
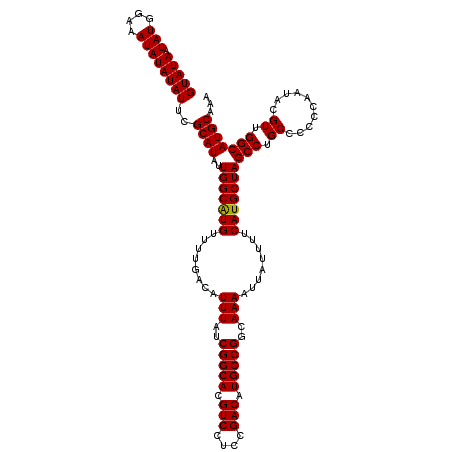
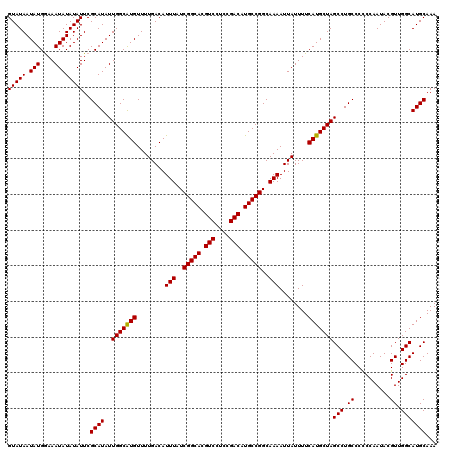
| Location | 4,054,651 – 4,054,771 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -31.08 |
| Energy contribution | -30.89 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4054651 120 + 20766785 GUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCGUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUAUUUUCAUGCUAGCCUGCCCCCCAAUACGUUGGCAUGCAAA (((((.(((....))))))))..((((..(((((((........(((..(((((.(((....))).)))))..)))........)))))))(((.((..........)).)))))))... ( -31.89) >DroSec_CAF1 37331 120 + 1 GUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUAUUUUCAUGCUAGCCUGCCCCCCAAUACGUUGGCAUGCAAA (((((.(((....))))))))..((((..(((((((........(((..(((((.(((....))).)))))..)))........)))))))(((.((..........)).)))))))... ( -32.29) >DroSim_CAF1 43457 120 + 1 GUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUAUUUUCAUGCUAGCCUGCCCCCCAAUACGUUGGCAUGCAAA (((((.(((....))))))))..((((..(((((((........(((..(((((.(((....))).)))))..)))........)))))))(((.((..........)).)))))))... ( -32.29) >DroEre_CAF1 37291 120 + 1 GUAUAAUAUGAAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCUGACAUGCCGGCAAAAUUAUUUUCAUGCUAGCCUGCCCUCCAAUACGUUGGCAUGCAAA ....((((((..(((....)))..))))))((((((........(((..(((((.(((....))).)))))..)))........)))))).((.((((..(......)..)))).))... ( -32.29) >consensus GUAUAAUAUGGAAAUAUAUAUUCGCAUAUUGGCAUGUUUUGACAUUUAUCGGCACGUCCUCCGACAUGCCGGCAAAAUUAUUUUCAUGCUAGCCUGCCCCCCAAUACGUUGGCAUGCAAA ....((((((......)))))).((((..(((((((........(((..(((((.(((....))).)))))..)))........)))))))(((.((..........)).)))))))... (-31.08 = -30.89 + -0.19)

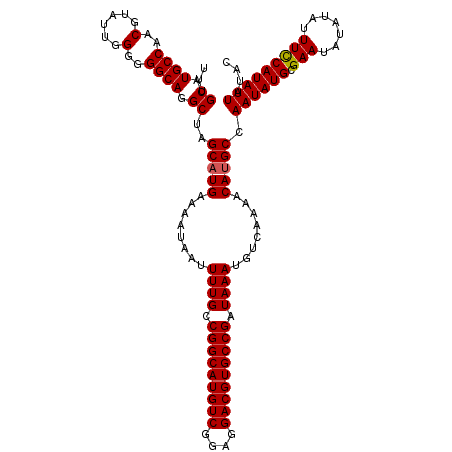
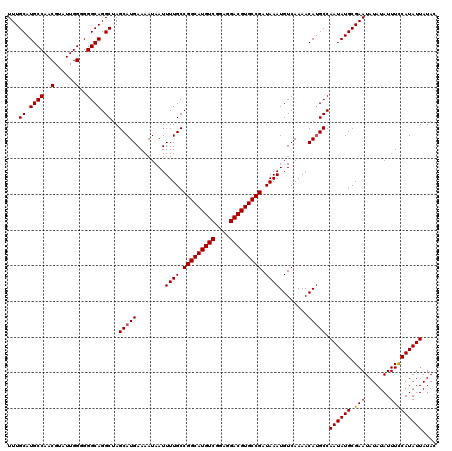
| Location | 4,054,651 – 4,054,771 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -36.05 |
| Energy contribution | -36.12 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4054651 120 - 20766785 UUUGCAUGCCAACGUAUUGGGGGGCAGGCUAGCAUGAAAAUAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACACGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUAC ((((((((((..(......)..))).(((..(((.((......)).)))(((((((((....)))))))))................)))...))))))).................... ( -34.80) >DroSec_CAF1 37331 120 - 1 UUUGCAUGCCAACGUAUUGGGGGGCAGGCUAGCAUGAAAAUAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUAC ...((.((((..(......)..)))).))..(((((........((((.(((((((((....))))))))).))))........))))).((((((.(((.......))))))))).... ( -38.09) >DroSim_CAF1 43457 120 - 1 UUUGCAUGCCAACGUAUUGGGGGGCAGGCUAGCAUGAAAAUAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUAC ...((.((((..(......)..)))).))..(((((........((((.(((((((((....))))))))).))))........))))).((((((.(((.......))))))))).... ( -38.09) >DroEre_CAF1 37291 120 - 1 UUUGCAUGCCAACGUAUUGGAGGGCAGGCUAGCAUGAAAAUAAUUUUGCCGGCAUGUCAGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUUCAUAUUAUAC ...((.((((..(......)..)))).))....(((((((((..((((((((((((((....)))))))))....((((....))))........)))))....)))))))))....... ( -35.50) >consensus UUUGCAUGCCAACGUAUUGGGGGGCAGGCUAGCAUGAAAAUAAUUUUGCCGGCAUGUCGGAGGACGUGCCGAUAAAUGUCAAAACAUGCCAAUAUGCGAAUAUAUAUUUCCAUAUUAUAC ...((.((((..(......)..)))).))..(((((........((((.(((((((((....))))))))).))))........))))).((((((.(((.......))))))))).... (-36.05 = -36.12 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:03 2006