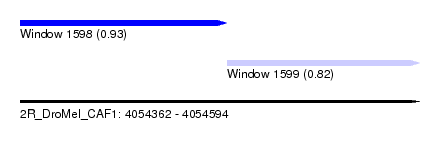
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,054,362 – 4,054,594 |
| Length | 232 |
| Max. P | 0.930959 |

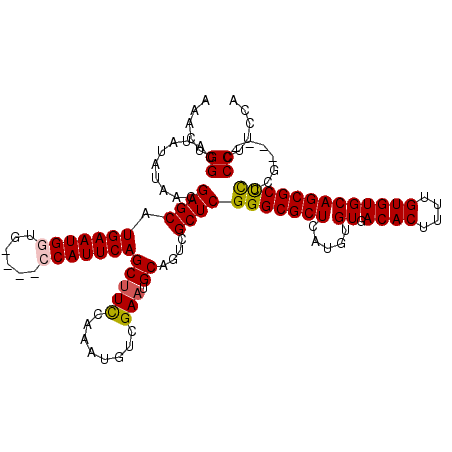
| Location | 4,054,362 – 4,054,482 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -31.88 |
| Energy contribution | -32.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4054362 120 + 20766785 AAGAAAUGUAAAUCGCGUUUUUGGCCCCAAGCUCUGCAAGCGCGUAGUUUCGAGAAGUGUCUGUUGAAUGGCCUUCAUGAGCUAGGUGCUUGUUAGCCGGAUUUUCAAGAAGCCAGACAU .((((((((.....))))))))(((.((..(((..((((((((.(((((.((.((((.(((........))))))).))))))).)))))))).))).)).(((....))))))...... ( -38.50) >DroSec_CAF1 37038 120 + 1 AAGAAAUGUAAAUCGCGUUUUUGGCCCCAAGCUCUGCAAGCGCGUAGUUCCGAGAAGUGUCUGUUGAAUGGCCUUCAUGAGCUAGGUGCUUGUUAGCCGGGUUUUCAAGAAGCCAGACAU .....((((.....((.((((((((((...(((..((((((((.((((((...((((.(((........)))))))..)))))).)))))))).))).))))...))))))))...)))) ( -43.00) >DroSim_CAF1 43164 120 + 1 AAGAAAUGUAAAUCGCGUUUUUGGCCCCAAGCUCUGCAAGCGCGUAGUUCCGAGAAGUGUCUGUUGAAUGGCCUUCAUGAGCUAGGUGCUUGUUAGCCGGGUUUUCAAGAAGCCAGACAU .....((((.....((.((((((((((...(((..((((((((.((((((...((((.(((........)))))))..)))))).)))))))).))).))))...))))))))...)))) ( -43.00) >DroEre_CAF1 36981 119 + 1 AAGCAAUGUAAAUCGCGUUUCUGGCCCAAAGCCCAGCA-GCGCGUAGUUCCGCGAAGUGUCUUUUGAAUGCGCUUUAUGAGCUAGGUGCUUGUUAGCCGGAUUUUCAAGAAGCCAGACAU ..((.((....)).))...((((((((...((..((((-((((.((((((...(((((((.........)))))))..)))))).)))).)))).)).)).(((....)))))))))... ( -33.70) >DroYak_CAF1 40470 120 + 1 CAGAAAUGUAAAUCGCGUUUCUGGUCCAAAGCCCUGCAAGCGCGUAGUUCUGCGAAGUGUCUUUUGAAUGGGAUUUAUGAGCUAGGUGCUUGUUAGCCGGGUUUUCAAGAAUCCAGACAU (((((((((.....)))))))))(((....((...((((((((.((((((....((((.((........)).))))..)))))).))))))))..)).((((((....)))))).))).. ( -44.00) >consensus AAGAAAUGUAAAUCGCGUUUUUGGCCCCAAGCUCUGCAAGCGCGUAGUUCCGAGAAGUGUCUGUUGAAUGGCCUUCAUGAGCUAGGUGCUUGUUAGCCGGGUUUUCAAGAAGCCAGACAU .((((((((.....))))))))(((..........((((((((.((((((...((((.(((........)))))))..)))))).))))))))..)))((.(((....))).))...... (-31.88 = -32.28 + 0.40)

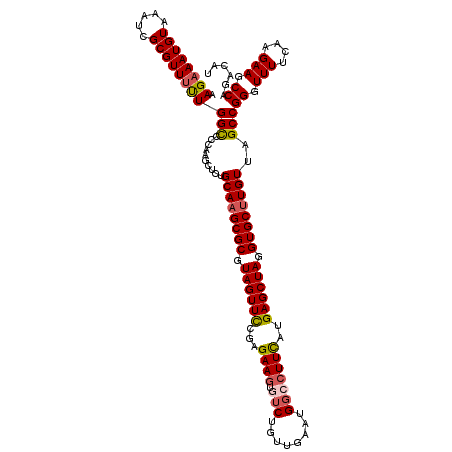
| Location | 4,054,482 – 4,054,594 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4054482 112 + 20766785 AAACUGGCAUAUAUAACGAGCAUGAAUGGUG----CCAUUCAGCUUCCAAAUGUCGAAUGCAGUCGCUCGGGCGCUCAUGUGUGACACUUUUGUGUGCAGCGCCCUCG----CCCUUCCA .....(((.........((((.((((((...----.))))))(((((........))).))....))))(((((((.....((.((((....)))))))))))))..)----))...... ( -38.40) >DroSec_CAF1 37158 116 + 1 AAACUGGCGUAUAUAACGAGCAUGAAUGGUG----CCAUUCAGCUUCCAAAUGUCGAAUGCAGUCGCUCGGGCGCUCAUGUGUGACACUUUUGUGUGCAGCGCCUUCAGUGCCCCUUCCA .....((..........((((.((((((...----.))))))(((((........))).))....))))(((((((...((((..(((......)))..))))....)))))))...)). ( -37.10) >DroSim_CAF1 43284 116 + 1 AAACUGGCAUAUAUAACGAGCAUGAAUGGUG----CCAUUCAGCUUCCAAAUGUCGAAUGCAGUCGCUCGGGCGCUCAUGUGUGACACUUUUGUGUGCAGCGCCUUCAGUGCCCCUUCCA .....(((((......(((((.((((((...----.))))))(((((........))).))....)))))((((((.....((.((((....))))))))))))....)))))....... ( -39.00) >DroEre_CAF1 37100 112 + 1 AAACUGGCAUAUAUAACGAGCAUGAAUGGUG----GCAUUCAGCUUUCAAAUGUCGAUUGCAGCCGCUCGGGCGCUCAUGUGUGACACUUUUGUGUGCAGCGCUCUCG----UCCUCACA .....(((.........((((.((((((...----.))))))((..((.......))..))....))))(((((((.....((.((((....)))))))))))))..)----))...... ( -32.20) >DroYak_CAF1 40590 116 + 1 AAACUGGCUUAUAUAACGAGCAUGAAUGCUGCCUGCCAUUCAGCUUUCGAAUGUCGAAUGCAGUCGCUCGGGCGCUCAUGUGUGACACUUUUGUGUGCAGCGCCCCCG----CCCUUCCA .....(((.........((((.(((((((.....).))))))((.((((.....)))).))....))))(((((((.....((.((((....)))))))))))))..)----))...... ( -40.40) >consensus AAACUGGCAUAUAUAACGAGCAUGAAUGGUG____CCAUUCAGCUUCCAAAUGUCGAAUGCAGUCGCUCGGGCGCUCAUGUGUGACACUUUUGUGUGCAGCGCCCUCG____CCCUUCCA .....((..........((((.(((((((......)))))))(((((........))).))....))))(((((((.....((.((((....)))))))))))))........))..... (-30.64 = -30.60 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:59 2006