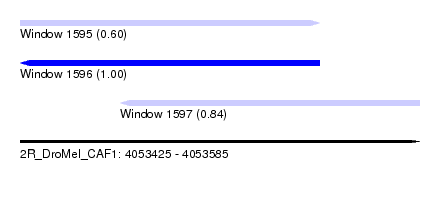
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,053,425 – 4,053,585 |
| Length | 160 |
| Max. P | 0.999018 |

| Location | 4,053,425 – 4,053,545 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -23.99 |
| Energy contribution | -25.47 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4053425 120 + 20766785 UGAAUGGAUUGCCUAUUGGUUCUGUGUUUGGAUUGGUUUUGCUGCACGUUGACUGGCAUGUAAAACAUGCCAAGAAAUCCAAAUAAAAUCCUUUUUGGAGCAGACAAGCCAUUGGUUCAG ......((..(((...((((((((((((((((((((((..((.....)).))))((((((.....))))))....)))))))))....(((.....))))))))...))))..))))).. ( -36.80) >DroSec_CAF1 36093 119 + 1 UGAAUGGAUUGCCUAUUGGUUCUGUGUUUG-UUUGGUUUUGCUGCACGUUAACUGGCAUGUAAAACAUGCCAAGAAAUCCAAAUAAAACCCUUUUCGGAGCAGACAAGCCAUUGGUUUAG ..........(((...(((((((((.((((-(((((((((((.....))....(((((((.....))))))).)))).)))))))))..((.....)).)))))...))))..))).... ( -31.40) >DroSim_CAF1 42218 120 + 1 UGAAUGGAUUGCCUAUUGGUUCUGUGUUUGGAUUGGUUUUGCUGCACGUUGACUGGCAUGUAAAACAUGCCAAGAAAUCCAAAUAAAACCCUUUUCGGAGCAGACAAGCCAUUGGUUUAG ..........(((...((((((((((((((((((((((..((.....)).))))((((((.....))))))....))))))))).....((.....)).)))))...))))..))).... ( -34.10) >DroEre_CAF1 36053 114 + 1 UGAAUGGAUUGCCUAUUGGUUCUGUGCUUGGAUUGGAUUGGU------UUGACUGGCAUGUAAAACAGGCCAAGAAAUCCAAAUAAAAUCCUUUUUGGAGCAGACAAGCCAUUGGUUGGG ..........(((...(((((((((..(((((((...(((((------(((((......))....))))))))..)))))))......(((.....))))))))...))))..))).... ( -31.70) >DroYak_CAF1 39544 109 + 1 UGAAUGGAUUGCCUAUUGGUUCUGUGCUUGGA-----UUGGU------UUGACUAGCAUGUAAAACAUGCCAAGAAAUUCAAAUAAAACCCUUUUGGGAGCAGACAAGCCAUUGGUGGAG ........(..((...(((((((((.((..((-----..(((------((.....(((((.....)))))...(.....).....)))))..))..)).)))))...))))..))..).. ( -30.80) >consensus UGAAUGGAUUGCCUAUUGGUUCUGUGUUUGGAUUGGUUUUGCUGCACGUUGACUGGCAUGUAAAACAUGCCAAGAAAUCCAAAUAAAACCCUUUUCGGAGCAGACAAGCCAUUGGUUGAG ..........(((...((((((((((((((((((...................(((((((.....)))))))...))))))))).....((.....)).)))))...))))..))).... (-23.99 = -25.47 + 1.48)

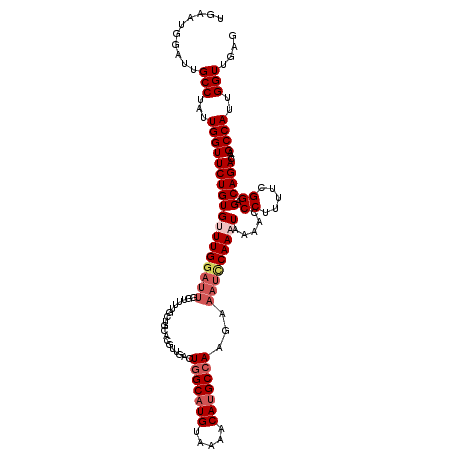
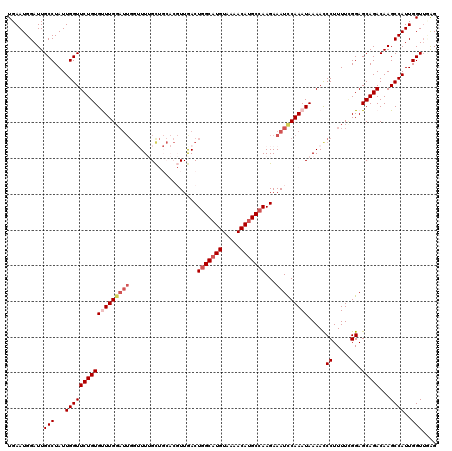
| Location | 4,053,425 – 4,053,545 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.06 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4053425 120 - 20766785 CUGAACCAAUGGCUUGUCUGCUCCAAAAAGGAUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUCAACGUGCAGCAAAACCAAUCCAAACACAGAACCAAUAGGCAAUCCAUUCA .......(((((.(((((((.(((.....))).....((((((((..((((((((.....)))))))).....((........)).))))))))...........))))))).))))).. ( -31.30) >DroSec_CAF1 36093 119 - 1 CUAAACCAAUGGCUUGUCUGCUCCGAAAAGGGUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUUAACGUGCAGCAAAACCAAA-CAAACACAGAACCAAUAGGCAAUCCAUUCA .......(((((.(((((((..((.....))((((..(((((......(((((((.....)))))))((......)).......)))))-.))))..........))))))).))))).. ( -29.90) >DroSim_CAF1 42218 120 - 1 CUAAACCAAUGGCUUGUCUGCUCCGAAAAGGGUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUCAACGUGCAGCAAAACCAAUCCAAACACAGAACCAAUAGGCAAUCCAUUCA .......(((((.(((((((((......))((((((.((((((((..((((((((.....)))))))).....((........)).))))))))...))))))..))))))).))))).. ( -33.50) >DroEre_CAF1 36053 114 - 1 CCCAACCAAUGGCUUGUCUGCUCCAAAAAGGAUUUUAUUUGGAUUUCUUGGCCUGUUUUACAUGCCAGUCAA------ACCAAUCCAAUCCAAGCACAGAACCAAUAGGCAAUCCAUUCA .......(((((.(((((((.........(((((....((((.((..(((((.((.....)).)))))..))------.))))...)))))..(........)..))))))).))))).. ( -25.50) >DroYak_CAF1 39544 109 - 1 CUCCACCAAUGGCUUGUCUGCUCCCAAAAGGGUUUUAUUUGAAUUUCUUGGCAUGUUUUACAUGCUAGUCAA------ACCAA-----UCCAAGCACAGAACCAAUAGGCAAUCCAUUCA .......(((((.(((((((..(((....))).....(((((.....((((((((.....))))))))))))------)....-----.................))))))).))))).. ( -25.50) >consensus CUAAACCAAUGGCUUGUCUGCUCCAAAAAGGGUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUCAACGUGCAGCAAAACCAAUCCAAACACAGAACCAAUAGGCAAUCCAUUCA .......(((((.(((((((.(((.....))).....((((((((...(((((((.....)))))))((.........))......))))))))...........))))))).))))).. (-24.74 = -25.06 + 0.32)

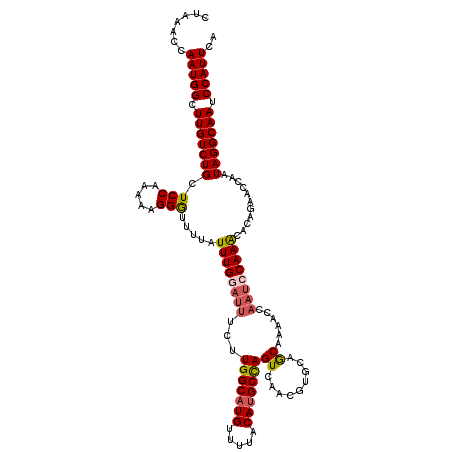
| Location | 4,053,465 – 4,053,585 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4053465 120 - 20766785 AAUUGAUAAAUGAAUUUCUAUACUGGCCGUCGCAUUCUUCCUGAACCAAUGGCUUGUCUGCUCCAAAAAGGAUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUCAACGUGCAGC .....................((.((((((.(..(((.....))).).)))))).))((((((((((..........))))))....((((((((.....)))))))).......)))). ( -27.60) >DroSec_CAF1 36132 120 - 1 AAUUGAUAAAUGAAUUUCUGUACUGGCAGCCGCAUUCUUCCUAAACCAAUGGCUUGUCUGCUCCGAAAAGGGUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUUAACGUGCAGC .................((((((.((((((((.................)))).))))...((((((..........))))))....((((((((.....)))))))).....)))))). ( -28.13) >DroSim_CAF1 42258 120 - 1 AAUUGAUAAAUGAAUUUCUGUACUGGCAGCCGCAUUCUUCCUAAACCAAUGGCUUGUCUGCUCCGAAAAGGGUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUCAACGUGCAGC .................((((((((((...........(((.(((((...(((......)))........))))).....))).....(((((((.....)))))))))))..)))))). ( -28.80) >DroEre_CAF1 36093 114 - 1 AAUUGAUAAAUGAAUUCCUGUACUGGCAGCAGCAUUCUUUCCCAACCAAUGGCUUGUCUGCUCCAAAAAGGAUUUUAUUUGGAUUUCUUGGCCUGUUUUACAUGCCAGUCAA------AC ..((((((((((((.((((....(((.(((((((..((............))..)).))))))))...)))).)))))))........((((.((.....)).)))))))))------.. ( -30.80) >DroYak_CAF1 39579 114 - 1 AAUUGAUAAAUGAAUUUCUGUACUGGCAGCAGCAUUCUUCCUCCACCAAUGGCUUGUCUGCUCCCAAAAGGGUUUUAUUUGAAUUUCUUGGCAUGUUUUACAUGCUAGUCAA------AC ..((((((((((((.((((....(((.(((((((..((............))..)).))))).)))..)))).)))))))........(((((((.....))))))))))))------.. ( -28.60) >consensus AAUUGAUAAAUGAAUUUCUGUACUGGCAGCCGCAUUCUUCCUAAACCAAUGGCUUGUCUGCUCCAAAAAGGGUUUUAUUUGGAUUUCUUGGCAUGUUUUACAUGCCAGUCAACGUGCAGC ..((((((((((((.((((.....(((((..(((..((............))..))))))))......)))).)))))))........(((((((.....))))))))))))........ (-24.42 = -24.10 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:57 2006