
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,050,938 – 4,051,044 |
| Length | 106 |
| Max. P | 0.999914 |

| Location | 4,050,938 – 4,051,044 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -15.40 |
| Energy contribution | -16.96 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4050938 106 + 20766785 UGCGGCCUUCGAUAUACUUGACGUAUUUUCUGAUACAUG---------GAUCAUGAAAAUCAGUUAAAGU-----GGCCGCAAUGAAAAUUAAUGCAUGGCUCAUGGUCAAUGCACUUAA (((((((........(((((((..((((((((((.....---------.)))).))))))..))).))))-----)))))))...........((((((((.....))).)))))..... ( -30.00) >DroSec_CAF1 33431 95 + 1 UGCGGCCUUCGUUAUACUUGACGUAUUUUCUGAUUCAUG---------GAUCAUGAAUA-----AAAAAU-----GGCCGCAA------UUAAUGCAUGGCUCAUGGUCAAUGCACUUAA (((((((..(((((....))))).(((((...(((((((---------...))))))).-----.)))))-----))))))).------....((((((((.....))).)))))..... ( -31.00) >DroSim_CAF1 38363 106 + 1 UGCGGCCUUCGUUAUACUUGACGUAUUUUCUGAUUCAUG---------GAUCAUGAAAAUCAGUUAAAAU-----GGCCGCAAUGAAAAUUAAUGCAUGGCUCAUGGUCAAUGCACUUAA (((((((..(((((....))))).(((((((((((....---------))))).))))))..........-----)))))))...........((((((((.....))).)))))..... ( -32.70) >DroEre_CAF1 32893 115 + 1 UGCGGCCUUCAC-----UUGACUUAUUUUCCGAUUCAUAUGAUUUUCUGAUCAUGAAAUUCAGUAAAAAAGAAAAAGCCGCAAUUUAAAUUAAUGCAUUGCUCACGGUCAAUGCAUUUCA ((((((.((..(-----((.(((...((((........((((((....))))))))))...)))....)))..)).)))))).........((((((((((.....).)))))))))... ( -26.60) >DroYak_CAF1 36836 90 + 1 UGCGACCUUCACUAUACUUGACUCAUUUUCUGAUU-------------------------CAGUAAAAUU-----GGCCGCAGGUAAAAUUAAUGCAUUGCUCAUGGUCAAUGCAUUUAA ((((.((.......((((.((.(((.....))).)-------------------------))))).....-----)).)))).........((((((((((.....).)))))))))... ( -20.50) >consensus UGCGGCCUUCGCUAUACUUGACGUAUUUUCUGAUUCAUG_________GAUCAUGAAAAUCAGUAAAAAU_____GGCCGCAAUGAAAAUUAAUGCAUGGCUCAUGGUCAAUGCACUUAA (((((((.(((.......)))............((((((............))))))..................)))))))...........((((((((.....))).)))))..... (-15.40 = -16.96 + 1.56)

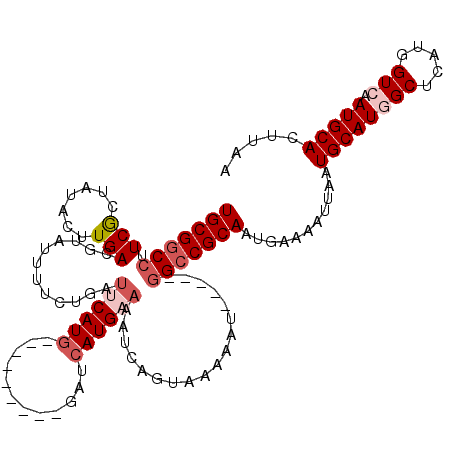
| Location | 4,050,938 – 4,051,044 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -17.18 |
| Energy contribution | -19.06 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.57 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4050938 106 - 20766785 UUAAGUGCAUUGACCAUGAGCCAUGCAUUAAUUUUCAUUGCGGCC-----ACUUUAACUGAUUUUCAUGAUC---------CAUGUAUCAGAAAAUACGUCAAGUAUAUCGAAGGCCGCA ...(((((((.(........).))))))).........(((((((-----.......(((((...((((...---------)))).)))))...((((.....))))......))))))) ( -29.30) >DroSec_CAF1 33431 95 - 1 UUAAGUGCAUUGACCAUGAGCCAUGCAUUAA------UUGCGGCC-----AUUUUU-----UAUUCAUGAUC---------CAUGAAUCAGAAAAUACGUCAAGUAUAACGAAGGCCGCA ...(((((((.(........).)))))))..------.(((((((-----((((((-----((((((((...---------))))))).))))))).(((........)))..))))))) ( -29.20) >DroSim_CAF1 38363 106 - 1 UUAAGUGCAUUGACCAUGAGCCAUGCAUUAAUUUUCAUUGCGGCC-----AUUUUAACUGAUUUUCAUGAUC---------CAUGAAUCAGAAAAUACGUCAAGUAUAACGAAGGCCGCA ...(((((((.(........).))))))).........(((((((-----.......(((((..(((((...---------))))))))))...((((.....))))......))))))) ( -29.90) >DroEre_CAF1 32893 115 - 1 UGAAAUGCAUUGACCGUGAGCAAUGCAUUAAUUUAAAUUGCGGCUUUUUCUUUUUUACUGAAUUUCAUGAUCAGAAAAUCAUAUGAAUCGGAAAAUAAGUCAA-----GUGAAGGCCGCA ...(((((((((..(....)))))))))).........((((((((((.(((.(((.((((.((..(((((......)))))..)).)))).))).)))....-----..)))))))))) ( -33.50) >DroYak_CAF1 36836 90 - 1 UUAAAUGCAUUGACCAUGAGCAAUGCAUUAAUUUUACCUGCGGCC-----AAUUUUACUG-------------------------AAUCAGAAAAUGAGUCAAGUAUAGUGAAGGUCGCA ...(((((((((........)))))))))..........((((((-----.(((.(((((-------------------------(.(((.....))).)).)))).)))...)))))). ( -27.50) >consensus UUAAGUGCAUUGACCAUGAGCCAUGCAUUAAUUUUAAUUGCGGCC_____AUUUUUACUGAUUUUCAUGAUC_________CAUGAAUCAGAAAAUACGUCAAGUAUAACGAAGGCCGCA ...(((((((.(........).))))))).........(((((((............((((..((((((............))))))))))...((((.....))))......))))))) (-17.18 = -19.06 + 1.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:54 2006