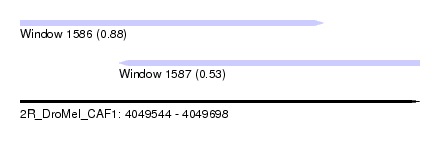
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,049,544 – 4,049,698 |
| Length | 154 |
| Max. P | 0.884555 |

| Location | 4,049,544 – 4,049,661 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.47 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.82 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4049544 117 + 20766785 --CUUAACUCGCUAAUAAAGUUAUGAAAAUGUUUGAGUUUGCAAACCCUU-UUUGUACGCUGUCAUCAGCCUGACAGUGCGAGCGGUUUGUCAUGAAUCUAAUUAGCGAGAGGACUCCCG --.....(((((((((....((((((...((((((....((((((.....-))))))((((((((......)))))))))))))).....)))))).....))))))))).((....)). ( -37.30) >DroSec_CAF1 32310 118 + 1 --CUUAACUCGCUAAUAAAGUUAUGAAAAUGUUUGAGUUUGCAAACCCUUUUUUGUACGCUGUCAUCAGCCUGACAGCGCGAGCGGUUUGUCAUGGAUCUAAUUAGCAAAAGGACUCCCG --........((((((....((((((...((((((....((((((......))))))((((((((......)))))))))))))).....)))))).....))))))....((....)). ( -32.10) >DroSim_CAF1 36981 118 + 1 --CUUAACUCGCUAAUAAAGUUAUGAAAAUGUUUGAGUUUGCAAACCCUUUUUUGUACGCUGUCAUCAGGCUGACAGCGCGAGCGGUUUGUCAUGGAUCUAAUUAGCGAAAGGACUCCCG --......((((((((....((((((...((((((....((((((......))))))((((((((......)))))))))))))).....)))))).....))))))))..((....)). ( -35.30) >DroEre_CAF1 31471 118 + 1 --CUCAACUCGCUAAUAAAGUUAUGAGAAUGUUUGAGAUUGCAAACCCUUUUUUGAGCGCUGUCGUCAGCCUGACAGCGCUAGCGGUUUAUCAAUGAUCUAAUUAGCGUGAGGACUCCUG --((((...(((((((.(..(((((((((((((((......))))).........((((((((((......))))))))))....)))).)).))))..).)))))))))))........ ( -33.80) >DroYak_CAF1 35342 114 + 1 CUCUUAGCUCGCUAAUAAAGUUAUGAAAAUGUUUGAGUUUUCAAACCCUUUUUUGAACGCUGUCGUCAGUCUGACAGCGCGAG------AUCAAGGAUCUAAUUACCCCAAGGACUCCUG .......(((((.......(((..((((..(((((......)))))..))))...)))(((((((......))))))))))))------....(((((((...........))).)))). ( -30.10) >consensus __CUUAACUCGCUAAUAAAGUUAUGAAAAUGUUUGAGUUUGCAAACCCUUUUUUGUACGCUGUCAUCAGCCUGACAGCGCGAGCGGUUUGUCAUGGAUCUAAUUAGCGAAAGGACUCCCG ..(((...((((((((....((((((...((((((....((((((......))))))((((((((......)))))))))))))).....)))))).....)))))))))))........ (-25.78 = -26.82 + 1.04)

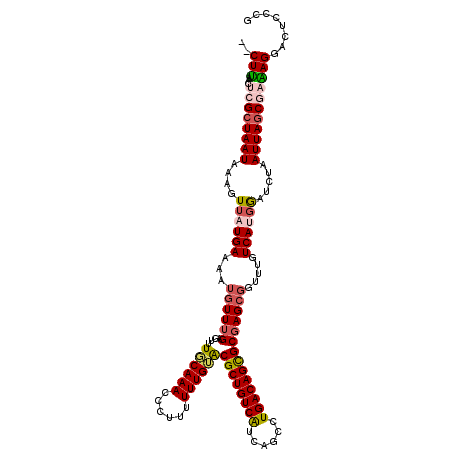
| Location | 4,049,582 – 4,049,698 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -24.24 |
| Energy contribution | -25.84 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4049582 116 - 20766785 ---CGUUUACAAUUUUGUCAUUCCCUGACUUGCAUUUCCGCGGGAGUCCUCUCGCUAAUUAGAUUCAUGACAAACCGCUCGCACUGUCAGGCUGAUGACAGCGUACAAA-AAGGGUUUGC ---............(((((((.((((((.(((......((((((....)))))).............((........)))))..))))))..)))))))(((.((...-....)).))) ( -30.20) >DroSec_CAF1 32348 117 - 1 ---UGUUUACAAUUUUGUCAUUCCCUGACUUGCAUUUCCGCGGGAGUCCUUUUGCUAAUUAGAUCCAUGACAAACCGCUCGCGCUGUCAGGCUGAUGACAGCGUACAAAAAAGGGUUUGC ---.............((((.....))))..(((..(((((((..(((......((....))......)))...))))..(((((((((......)))))))))........)))..))) ( -34.40) >DroSim_CAF1 37019 117 - 1 ---UGUUUACAAUUUUGUCAUUCCCUGACUUGCAUUUCCGCGGGAGUCCUUUCGCUAAUUAGAUCCAUGACAAACCGCUCGCGCUGUCAGCCUGAUGACAGCGUACAAAAAAGGGUUUGC ---.............((((.....))))..(((..(((((((..(((...((........)).....)))...))))..(((((((((......)))))))))........)))..))) ( -34.70) >DroEre_CAF1 31509 120 - 1 GCAAUUUUGUAAUUUUGUCAUUCUCUGACUUGCACUUCCGCAGGAGUCCUCACGCUAAUUAGAUCAUUGAUAAACCGCUAGCGCUGUCAGGCUGACGACAGCGCUCAAAAAAGGGUUUGC ((((..........((((((...((((((((((......)))))(((......)))..)))))....))))))(((.(((((((((((........)))))))))......))))))))) ( -32.90) >DroYak_CAF1 35382 110 - 1 ---UAUUUGCAGUUUUAUCAUU-UCUGACUUGCAUUUCCGCAGGAGUCCUUGGGGUAAUUAGAUCCUUGAU------CUCGCGCUGUCAGACUGACGACAGCGUUCAAAAAAGGGUUUGA ---....(((((((........-...))).))))...((.((((....)))).))...((((((((((...------...((((((((........))))))))......)))))))))) ( -33.50) >consensus ___UGUUUACAAUUUUGUCAUUCCCUGACUUGCAUUUCCGCGGGAGUCCUUUCGCUAAUUAGAUCCAUGACAAACCGCUCGCGCUGUCAGGCUGAUGACAGCGUACAAAAAAGGGUUUGC ................((((.....))))..(((..(((((((..(((......((....))......)))...))))..(((((((((......)))))))))........)))..))) (-24.24 = -25.84 + 1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:47 2006