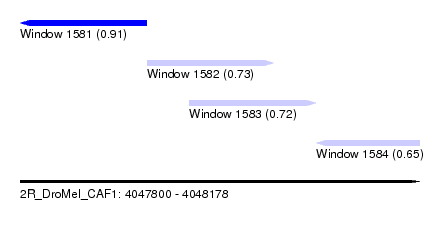
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,047,800 – 4,048,178 |
| Length | 378 |
| Max. P | 0.911833 |

| Location | 4,047,800 – 4,047,920 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -26.22 |
| Energy contribution | -28.62 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047800 120 - 20766785 GGAAUUGCAUAUUUAGCAGAACAUUGAAAAUCAGGAACCGGCAAGUGCGGAAACCUGCGCUGUUGGUCUUCAUUUGCAUAUUGCUGAUUGAGUAUGAAUUUUGGCCAGCAGUUGCAUAGA ((((((.((((((((((((.....((.((((.((((.((((((.((((((....)))))))))))))))).)))).)).....))).))))))))))))))).....((....))..... ( -36.40) >DroSec_CAF1 30544 120 - 1 GGAAUUGCAUAUUUAGCAGAACAUUGAAAAUCAGGGACCGGCAAGUGCGGAAACCUGCGCUGUUGGUCUUCAUUUGCAUAUUGCUGAUUGAGUAUGAAUUUUGGCAAGCAGUUGCAUAGA (.((((((.......(((((...(((.....)))(((((((((.((((((....)))))))))))))))...)))))...((((..(.............)..)))))))))).)..... ( -39.82) >DroSim_CAF1 35213 120 - 1 GCAAUUGCAUAUUUAGCAGAACAUUGAAAAUCAGGGACCGGCAAGUGCGGAAACCUGCGCUGUUGGUCUUCAUUUGCAUAUUGCUGAUUGAGUAUGAAUUUUGGCAAGCAGUUGCAUAGA ((((((((.......(((((...(((.....)))(((((((((.((((((....)))))))))))))))...)))))...((((..(.............)..))))))))))))..... ( -44.32) >DroEre_CAF1 29729 106 - 1 GGAAUUGCAUAUUUAGCAGAACACUGAAAAUCAGU--------------GAAACCUGCGCUGUUGGUCUUCAUUUGAAUAUUGCUGAUUGAGUAUGAAUUUUGGCAAGCAGUUGCUUGUA ((((((.((((((((((((..(((((.....))))--------------)....))))...((..((.(((....)))....))..)))))))))))))))).((((((....)))))). ( -29.50) >DroYak_CAF1 33526 120 - 1 GGAAUUGCAUAUUUAGCAGAACAUUGAAAAUCAGUGGGCGGUUGGUGGAGAAACCUGCGCUAUUGCUCCUCAUUUGCAUAUUGCUGAUUGAGUAUGAAUUUUGGCAAGCAGUUGCUUAUA ((((((.((((((((((((.....((.((((.((.(((((((.((((.((....)).))))))))))))).)))).)).....))).)))))))))))))))...((((....))))... ( -33.80) >consensus GGAAUUGCAUAUUUAGCAGAACAUUGAAAAUCAGGGACCGGCAAGUGCGGAAACCUGCGCUGUUGGUCUUCAUUUGCAUAUUGCUGAUUGAGUAUGAAUUUUGGCAAGCAGUUGCAUAGA (.((((((.......(((((...(((.....))).(((((((.(((((((....))))))))))))))....)))))...((((..(.............)..)))))))))).)..... (-26.22 = -28.62 + 2.40)

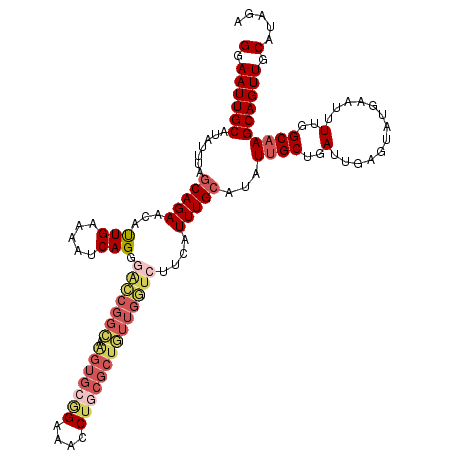
| Location | 4,047,920 – 4,048,040 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -31.18 |
| Energy contribution | -31.34 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047920 120 + 20766785 UUCAAAUGAAAUAAAACAGUACGACAGCGAGUUAAGUUGCUCUAUCCGUUGUUUUGCGCGGAUUUGGCGUGCUUCCUGCUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCGCAAAACGU ..................(((.((((((((((......)))).....)))))).)))((((((((.(((..(....((((((.......))))))...)..))).))))))))....... ( -37.20) >DroSec_CAF1 30664 120 + 1 UUCAAAUGAAAUAAAACAGUACGACAGCGAGUUAAGUUGCUCUAUCCGUUGUUUUGUGCGGAUUUGGCGUGCUUUCUGCUAUUUUCCCCGUGGCAAACGUGCGCGAAAUUCGCAAACCGU ..........((((((((..(((.....((((......))))....)))))))))))((((((((.(((..(.((.((((((.......)))))))).)..))).))))))))....... ( -36.20) >DroSim_CAF1 35333 120 + 1 UUCAAAUGAAAUAAAACAGUACGACAGCGAGUUAAGUUGCUCUAUCCGUUGUUUUGCGCGGAUUUGGCGUGCUUCCUGCUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCGCAAACCGU ..................(((.((((((((((......)))).....)))))).)))((((((((.(((..(....((((((.......))))))...)..))).))))))))....... ( -37.20) >DroEre_CAF1 29835 120 + 1 UCCAAAUGAAAUAAAACAGCGCGACAGCGAGUUAAGUUCCUCUAUCCGUUGUUUUGCGCGGAUCUCGCGUGCGUCCUGCUAUUUUGCCCGUGGCAAACGUGCGCGGAAUUCGCAAACCGU ...........(((((((((((....))(((........))).....))))))))).((((((..((((..(((..((((((.......)))))).)))..))))..))))))....... ( -43.70) >DroYak_CAF1 33646 120 + 1 UUCAAAUGAAAUAAAAGAGUACGACAGCGAGUUAAGUUUCCCUAUCCGUUGUUUUGCGCGGAUUUAGCGUGCUUUCUGAUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCAAAAACGGU ......((((.....((((((((.(((.(((......))).))((((((........))))))...)))))))))............(((((.((....)))))))..))))........ ( -25.10) >consensus UUCAAAUGAAAUAAAACAGUACGACAGCGAGUUAAGUUGCUCUAUCCGUUGUUUUGCGCGGAUUUGGCGUGCUUCCUGCUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCGCAAACCGU ..................(((.(((((((.(...((.....))..)))))))).)))((((((((.(((..(....((((((.......))))))...)..))).))))))))....... (-31.18 = -31.34 + 0.16)

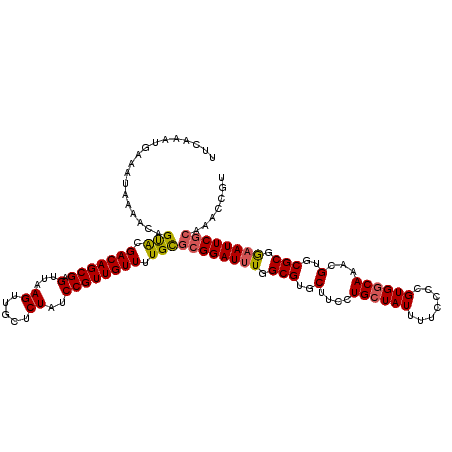
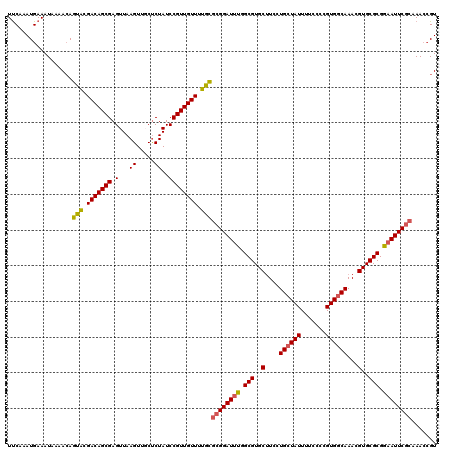
| Location | 4,047,960 – 4,048,080 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -28.54 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047960 120 + 20766785 UCUAUCCGUUGUUUUGCGCGGAUUUGGCGUGCUUCCUGCUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCGCAAAACGUAAAAAAUUCAUUUCAUCCAUUUCGAAAUAGUAGGAAGCGA ...((((((........))))))......((((((((((((((((....((((...((((..(((.....)))...))))................))))...)))))))))))))))). ( -35.31) >DroSec_CAF1 30704 120 + 1 UCUAUCCGUUGUUUUGUGCGGAUUUGGCGUGCUUUCUGCUAUUUUCCCCGUGGCAAACGUGCGCGAAAUUCGCAAACCGUAAAAAAUUGAUUUCAUCCAUUUCGUAAUAGCAGGAAGCGA ...((((((........))))))......((((((((((((((......((((((....)))(((.....)))........................))).....)))))))))))))). ( -33.70) >DroSim_CAF1 35373 120 + 1 UCUAUCCGUUGUUUUGCGCGGAUUUGGCGUGCUUCCUGCUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCGCAAACCGUAAAAAAUUCAUUUCAUCCAUUUCGUAAUAGCAGGAAGCGA ...((((((........))))))......((((((((((((((......((((((....)))(((.....)))........................))).....)))))))))))))). ( -35.00) >DroEre_CAF1 29875 119 + 1 UCUAUCCGUUGUUUUGCGCGGAUCUCGCGUGCGUCCUGCUAUUUUGCCCGUGGCAAACGUGCGCGGAAUUCGCAAACCGUAAAAAAUUCAU-UCAUCCAUUUCGGAAUAGUAGAAAGCGA ......((((.((((((((((((..((((..(((..((((((.......)))))).)))..))))..))))))..................-...(((.....)))...)))))))))). ( -41.30) >DroYak_CAF1 33686 120 + 1 CCUAUCCGUUGUUUUGCGCGGAUUUAGCGUGCUUUCUGAUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCAAAAACGGUAAAAAAUUCAUUUCAUUCAUUUCGGAAUAGUAGGAAGCGA ...((((((........))))))......(((((((((.....(((((((((.((....)))))))...........((........))..............))))...))))))))). ( -26.80) >consensus UCUAUCCGUUGUUUUGCGCGGAUUUGGCGUGCUUCCUGCUAUUUUCCCCGUGGCAAACGUGCGCGGAAUUCGCAAACCGUAAAAAAUUCAUUUCAUCCAUUUCGGAAUAGUAGGAAGCGA ...((((((........))))))......((((((((((((((((....((((((....)))(((.....)))........................)))...)))))))))))))))). (-28.54 = -29.46 + 0.92)

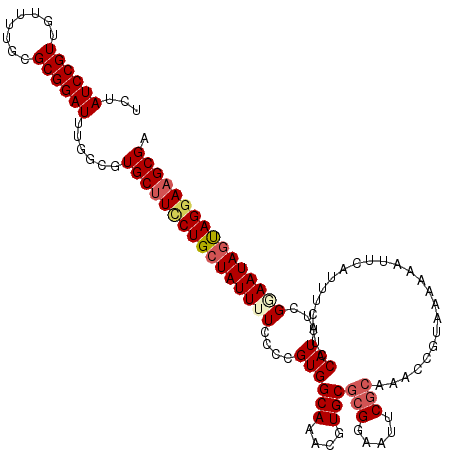
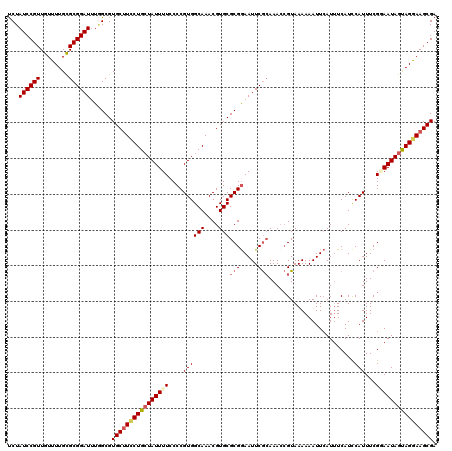
| Location | 4,048,080 – 4,048,178 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -26.89 |
| Energy contribution | -27.45 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4048080 98 - 20766785 -------------------UUGGUCAGCGGCGCUUUUUAUGCAAAUAACAAGCUGCCAGACAAUCGGGCUUGACGUGAGGACUUUAAGCCGCAUACCGCCGCGAUUUGCCGUUUAAA -------------------..(((..(((((((((.((((....)))).))))(((..(.....).(((((((.((....)).))))))))))....))))).....)))....... ( -29.90) >DroSec_CAF1 30824 117 - 1 UAAGCCCAACAUGCUCCAGUUGGUCAGCGGCGCUUUUUAUGCAAAUAACAAGCUGCCAGACAAUCGGGCUUGAAGUGAGGACUUUAAGCCGCAUACCGCCGCACUUUGCCGUUUAAA ((((((((((........)))))...(((((((((.((((....)))).))))(((..(.....).((((((((((....)))))))))))))....)))))........))))).. ( -39.10) >DroSim_CAF1 35493 117 - 1 UAAGCCCAACAUGCUCCAGUUGGUCAGCGGCGCUUUUUAUGCAAAUAACAAACUGCCAGACAAUCGGGCUUGAAGUGAGGACUUUAAGCCGCAUACCGCCGCACUUUGCCGUUUAAA ((((((((((........)))))...((((((.....(((((................(.....).((((((((((....))))))))))))))).))))))........))))).. ( -37.20) >DroYak_CAF1 33806 115 - 1 UAAGCCCAAUGUGCUACAGUUGGUCAGCGGCGCU--UUAUGCAAAUAACAAGCUGCCAGACAAUCAAGCUUGAAGUGAGGACUUUAAGCCGCAUACCGCCGGAGUUUGCUGUUUAAG (((((((..(((((........(((.(((((...--((((....))))...)))))..)))......(((((((((....))))))))).))))).....)).)))))......... ( -30.20) >consensus UAAGCCCAACAUGCUCCAGUUGGUCAGCGGCGCUUUUUAUGCAAAUAACAAGCUGCCAGACAAUCGGGCUUGAAGUGAGGACUUUAAGCCGCAUACCGCCGCACUUUGCCGUUUAAA .....................(((..((((((.....(((((................(.....).((((((((((....))))))))))))))).)))))).....)))....... (-26.89 = -27.45 + 0.56)

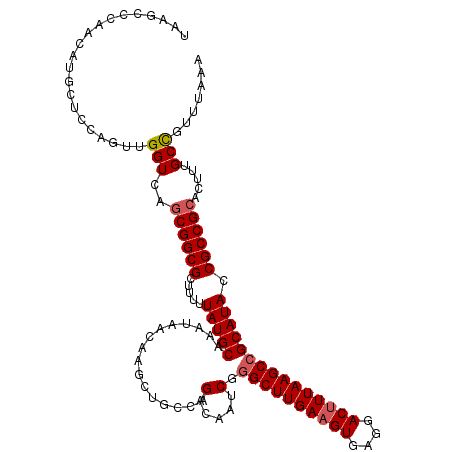
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:43 2006