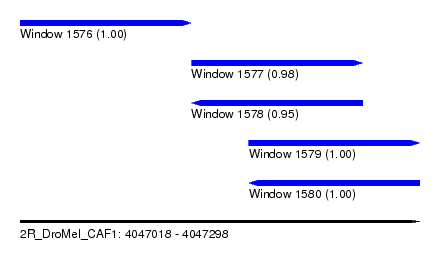
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,047,018 – 4,047,298 |
| Length | 280 |
| Max. P | 0.999411 |

| Location | 4,047,018 – 4,047,138 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -40.82 |
| Energy contribution | -40.62 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047018 120 + 20766785 UUUUGGCAAGUGGGAAACUUUGUCCUGCUGCCAUGACAAUAAUCGUGGUAAUGGGCUGUCUCAUAAUAAUGCUAAUGAUGCCUCUGGGUGCGGGUCUGGACCCGACACGCCCAGCGAAUU ..((((((.((((((......((((...((((((((......))))))))..))))..)))))).....))))))......(.((((((((((((....)))))...))))))).).... ( -44.20) >DroSec_CAF1 29768 120 + 1 UUUUGGCAAGUGGGAAACUUUGUCCUGCUGCCAUGACAAUAAUCGUGGUAAUGGGCUGUCUCAUAAUAAUGCUAAUGAUGCCUCUGGGUACGGGUCUGGACCCGACACGCCCAGCGAAUU ..((.((.((..(((.......)))..))(((((((......)))))))..(((((((((..................((((....)))).((((....)))))))).))))))).)).. ( -42.10) >DroSim_CAF1 34439 120 + 1 UUUUGGCAAGUGGGAAACUUUGUCCUGCUGCCAUGACAAUAAUCGUGGUAAUGGGCUGUCUCAUAAUAAUGCUAAUGAUGCCUCUGGGUACGGGUCUGGACCCGACACGCCCAGCGAAUU ..((.((.((..(((.......)))..))(((((((......)))))))..(((((((((..................((((....)))).((((....)))))))).))))))).)).. ( -42.10) >DroEre_CAF1 29112 120 + 1 UUUUGGCAAGUGGGAAACUUUGUCCUGCUGCCAUGACAAUAAUCGUGGUAAUGGGCUGUCUCAUAAUAAUGCUAAUGAUGCCUCUGGGUACGGGUCUGGACCCGACACGCCCAGCAAAUU ....(((((..((....))))))).(((((((((((......)))))))).(((((((((..................((((....)))).((((....)))))))).)))))))).... ( -42.10) >DroYak_CAF1 32907 120 + 1 UUUUGCCAAGUGGGAAACUUUGUCCUGCUGCCAUGACAAUAAUCGUGGUAAUGGGCUGUCUCAUAAUAAUGCUAAUGAUGCCUCUGGGUACGGGUCUGGACCCGACACGCCCAGCAAAUU .(((((..((..(((.......)))..))(((((((......)))))))..(((((((((..................((((....)))).((((....)))))))).)))))))))).. ( -43.20) >consensus UUUUGGCAAGUGGGAAACUUUGUCCUGCUGCCAUGACAAUAAUCGUGGUAAUGGGCUGUCUCAUAAUAAUGCUAAUGAUGCCUCUGGGUACGGGUCUGGACCCGACACGCCCAGCGAAUU ....(((((..((....))))))).(((((((((((......)))))))).(((((((((..................((((....)))).((((....)))))))).)))))))).... (-40.82 = -40.62 + -0.20)

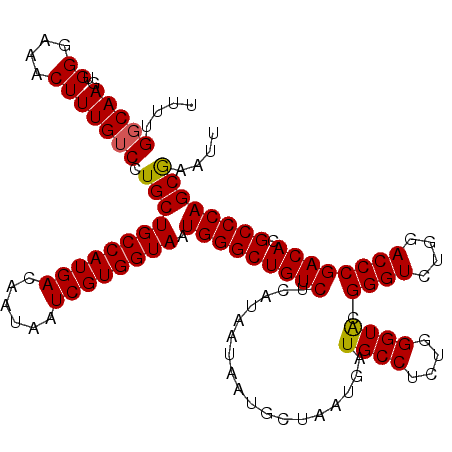
| Location | 4,047,138 – 4,047,258 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -33.20 |
| Energy contribution | -33.28 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047138 120 + 20766785 AUAACUGCCUCCGGAGCAAUGGCGAAAUUAGAGUUUCAAUUUCCUCUGCGAAUCCGGCUGCCGAAGGCGGGGAAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUU ......((((.((((((...((......(((((..........))))).....)).))).))).))))((((((((..((((((......)))))).))))))))............... ( -38.70) >DroSec_CAF1 29888 120 + 1 AUAACUGCCUCCGGUGCGAUGGUGAAAUUAGAGUUUCAAUUUCCUCUGCGAAUCCGGCAGCCAAAGGCGGGGCAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUU ....(((((.....((((..((((((((....))))))....))..)))).....)))))...(((((((((.(((..((((((......)))))).))).)))).((....)).))))) ( -35.40) >DroSim_CAF1 34559 120 + 1 AUAACUGCCUCCGGCGCGAUGGUGAAAUUAGAGUUUCAAUUUCCUCUGCGAAUCCGGCUGCCAAAGGCGGGGCAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUU ......((..((((((((..((((((((....))))))....))..))))...))))..))..(((((((((.(((..((((((......)))))).))).)))).((....)).))))) ( -35.50) >DroEre_CAF1 29232 120 + 1 AUAACAGCCUCCGGCGCAAUGGCGAAAUUAGAGUUUCAAUUUCCGCAGCGAAUCCGGCUGCCAAAAGCGGGGAAAGCGGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUU ............((((.....(((((((....))))).......(((((.......))))).....))((((((((..((((((......)))))).)))))))).........)))).. ( -39.70) >DroYak_CAF1 33027 120 + 1 AUGACUGCCUCCGGCGCAAUGGCGAAAUUAGAGUUUCAAUUUCCGCAGCGAAUCCGGCUGCCAAAGGCGGGGAAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUU .(((..((((..(((((....((((((((.(.....)))))).)))..((....)))).)))..))))((((((((..((((((......)))))).))))))))....)))........ ( -39.20) >consensus AUAACUGCCUCCGGCGCAAUGGCGAAAUUAGAGUUUCAAUUUCCUCUGCGAAUCCGGCUGCCAAAGGCGGGGAAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUU ............(((((..(((((((((....)))))..........((.......)).))))...))((((((((..((((((......)))))).))))))))..........))).. (-33.20 = -33.28 + 0.08)

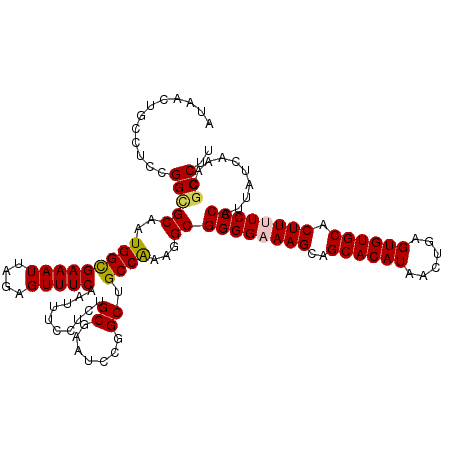
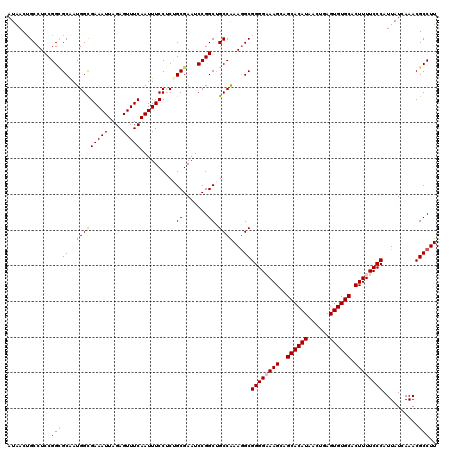
| Location | 4,047,138 – 4,047,258 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -35.62 |
| Energy contribution | -35.94 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047138 120 - 20766785 AAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUUCCCCGCCUUCGGCAGCCGGAUUCGCAGAGGAAAUUGAAACUCUAAUUUCGCCAUUGCUCCGGAGGCAGUUAU ..(((..........(((.((((((((((........))))).))))).)))((((((((.(((.((......((((..........)))).......))...))))))))))).))).. ( -39.92) >DroSec_CAF1 29888 120 - 1 AAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUGCCCCGCCUUUGGCUGCCGGAUUCGCAGAGGAAAUUGAAACUCUAAUUUCACCAUCGCACCGGAGGCAGUUAU ((((((((....)))(((..(((((((((........))))).))))..))))))))((((((((......((.((.(((((((......)))))))....))))......)))))))). ( -38.10) >DroSim_CAF1 34559 120 - 1 AAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUGCCCCGCCUUUGGCAGCCGGAUUCGCAGAGGAAAUUGAAACUCUAAUUUCACCAUCGCGCCGGAGGCAGUUAU ..(((..........(((..(((((((((........))))).))))..)))(((((((((.((.......)).(..(((((((......)))))))..).....))))))))).))).. ( -39.30) >DroEre_CAF1 29232 120 - 1 AAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCCGCUUUCCCCGCUUUUGGCAGCCGGAUUCGCUGCGGAAAUUGAAACUCUAAUUUCGCCAUUGCGCCGGAGGCUGUUAU ((((((.........((((((.(((((((........)))).)))))))))))))))((((((((.....(((.(((.((((((......)))))))))....))).....)))))))). ( -39.00) >DroYak_CAF1 33027 120 - 1 AAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUUCCCCGCCUUUGGCAGCCGGAUUCGCUGCGGAAAUUGAAACUCUAAUUUCGCCAUUGCGCCGGAGGCAGUCAU ..(((..........(((.((((((((((........))))).))))).)))((((((((((((.......)))(((.((((((......)))))))))......))))))))).))).. ( -42.90) >consensus AAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUUCCCCGCCUUUGGCAGCCGGAUUCGCAGAGGAAAUUGAAACUCUAAUUUCGCCAUUGCGCCGGAGGCAGUUAU ..(((..........(((.((((((((((........))))).))))).)))(((((((((.((.......))....(((((((......)))))))........))))))))).))).. (-35.62 = -35.94 + 0.32)

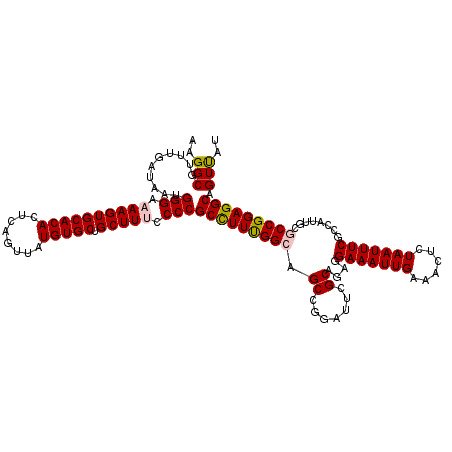
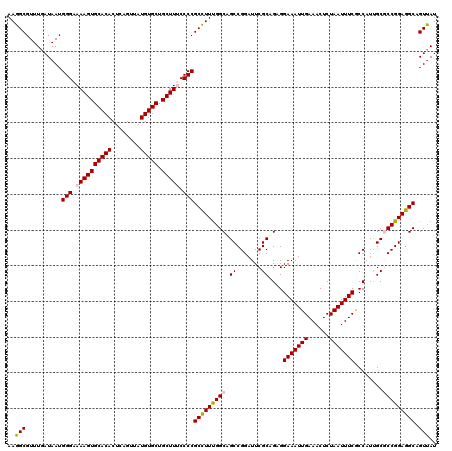
| Location | 4,047,178 – 4,047,298 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047178 120 + 20766785 UUCCUCUGCGAAUCCGGCUGCCGAAGGCGGGGAAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUUAAAGCGAAAACUUUUUAAUUGUGCCACAACAACCGGAGCU .......((...(((((.((.....(((((((((((..((((((......)))))).)))))))).........(((......)))................)))....)).))))))). ( -36.70) >DroSec_CAF1 29928 120 + 1 UUCCUCUGCGAAUCCGGCAGCCAAAGGCGGGGCAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUUAAAGCGAAAACUUUUUAAUUGUGCCACAACAACCGGAGCU ((((.((((.......)))).....(((((((.(((..((((((......)))))).))).)))).........(((......)))................))).........)))).. ( -31.30) >DroSim_CAF1 34599 120 + 1 UUCCUCUGCGAAUCCGGCUGCCAAAGGCGGGGCAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUUAAAGCGAAAACUUUUUAAUUGUGCCACAACAACCGGAGCU .......((...(((((.((.....(((((((.(((..((((((......)))))).))).)))).........(((......)))................)))....)).))))))). ( -32.80) >DroEre_CAF1 29272 120 + 1 UUCCGCAGCGAAUCCGGCUGCCAAAAGCGGGGAAAGCGGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUUAAAGCGAAAACUUUUUAAUUGUGCCACAACAAGCGGAGCU ((((((.((......(((.((.....))((((((((..((((((......)))))).))))))))..........))).....)).............((((......)))))))))).. ( -38.60) >DroYak_CAF1 33067 120 + 1 UUCCGCAGCGAAUCCGGCUGCCAAAGGCGGGGAAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUUAAAGCAAAAACUUUUUAAUUGUGCCACAACAACCGGAGCU (((((((((.......)))))....(((((((((((..((((((......)))))).))))))))..........((......)).................))).........)))).. ( -37.70) >consensus UUCCUCUGCGAAUCCGGCUGCCAAAGGCGGGGAAAGCAGCACAUAACUGAGUGUGCACUUUUCCCAUUAUCAAACGCCUUAAAGCGAAAACUUUUUAAUUGUGCCACAACAACCGGAGCU ((((...((......(((.((.....))((((((((..((((((......)))))).))))))))..........))).....)).............((((......))))..)))).. (-32.22 = -32.62 + 0.40)

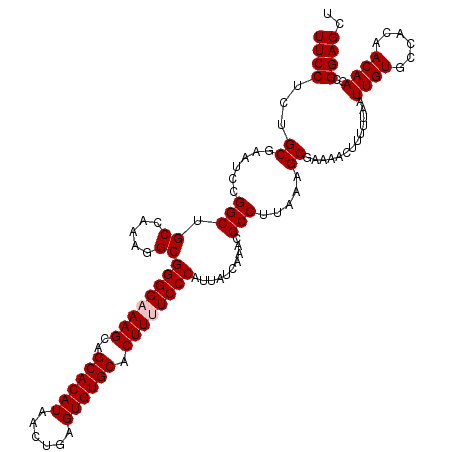
| Location | 4,047,178 – 4,047,298 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -40.94 |
| Consensus MFE | -40.20 |
| Energy contribution | -40.52 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4047178 120 - 20766785 AGCUCCGGUUGUUGUGGCACAAUUAAAAAGUUUUCGCUUUAAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUUCCCCGCCUUCGGCAGCCGGAUUCGCAGAGGAA .(((((((((((((.(((...........(((..((((....))))...)))...(((.((((((((((........))))).))))).))))))..)))))))))))...))....... ( -45.50) >DroSec_CAF1 29928 120 - 1 AGCUCCGGUUGUUGUGGCACAAUUAAAAAGUUUUCGCUUUAAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUGCCCCGCCUUUGGCUGCCGGAUUCGCAGAGGAA .((((((((.((((.(((...........(((..((((....))))...)))...(((..(((((((((........))))).))))..))))))..)))).))))))...))....... ( -38.60) >DroSim_CAF1 34599 120 - 1 AGCUCCGGUUGUUGUGGCACAAUUAAAAAGUUUUCGCUUUAAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUGCCCCGCCUUUGGCAGCCGGAUUCGCAGAGGAA .(((((((((((((.(((...........(((..((((....))))...)))...(((..(((((((((........))))).))))..))))))..)))))))))))...))....... ( -42.40) >DroEre_CAF1 29272 120 - 1 AGCUCCGCUUGUUGUGGCACAAUUAAAAAGUUUUCGCUUUAAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCCGCUUUCCCCGCUUUUGGCAGCCGGAUUCGCUGCGGAA ...(((((.((.(.((((...........(((..((((....))))...)))...(((.((((((((((........)))).)))))).)))((.....)).)))).)..))..))))). ( -36.20) >DroYak_CAF1 33067 120 - 1 AGCUCCGGUUGUUGUGGCACAAUUAAAAAGUUUUUGCUUUAAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUUCCCCGCCUUUGGCAGCCGGAUUCGCUGCGGAA ((((((((((((((.(((...((((((..(((((......))))).))))))...(((.((((((((((........))))).))))).))))))..)))))))))))...)))...... ( -42.00) >consensus AGCUCCGGUUGUUGUGGCACAAUUAAAAAGUUUUCGCUUUAAGGCGUUUGAUAAUGGGAAAAGUGCACACUCAGUUAUGUGCUGCUUUCCCCGCCUUUGGCAGCCGGAUUCGCAGAGGAA .(((((((((((((.(((...........(((..((((....))))...)))...(((.((((((((((........))))).))))).))))))..)))))))))))...))....... (-40.20 = -40.52 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:39 2006