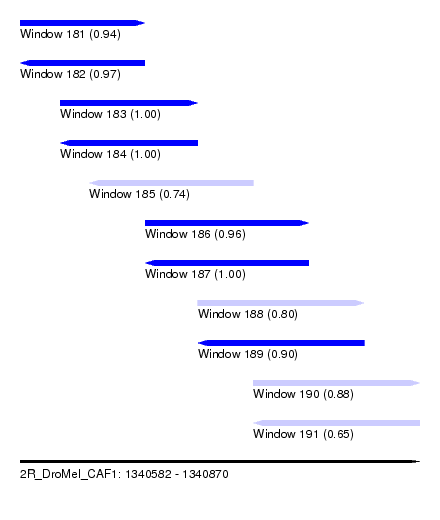
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,340,582 – 1,340,870 |
| Length | 288 |
| Max. P | 0.998948 |

| Location | 1,340,582 – 1,340,672 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -29.10 |
| Energy contribution | -29.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.37 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340582 90 + 20766785 CAUUCCUGCCACAUCCCCACUUCAGUCCUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAU ................................((((((((((((((((((..........)))))))....)))))))))))........ ( -29.10) >DroSec_CAF1 58994 90 + 1 CAUUCCGGCCAUGUCCCCACUUCGGUCCUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAU ......((((..((....))...)))).....((((((((((((((((((..........)))))))....)))))))))))........ ( -32.60) >DroYak_CAF1 27597 90 + 1 CAUUUCUGCUAUAUUACCACUUCGGUUCUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAU ..........(((..(((.....)))..))).((((((((((((((((((..........)))))))....)))))))))))........ ( -31.10) >consensus CAUUCCUGCCAUAUCCCCACUUCGGUCCUGUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAU ................................((((((((((((((((((..........)))))))....)))))))))))........ (-29.10 = -29.10 + -0.00)

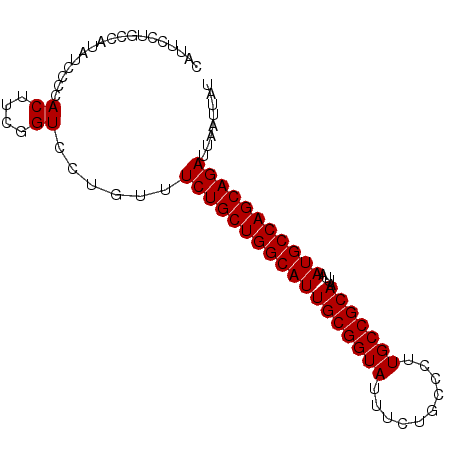
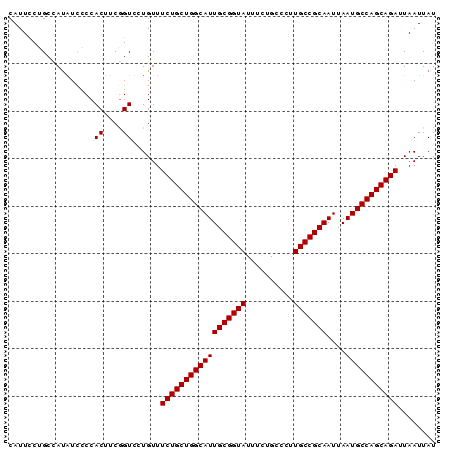
| Location | 1,340,582 – 1,340,672 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.34 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340582 90 - 20766785 AUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAGGACUGAAGUGGGGAUGUGGCAGGAAUG ........(((((((((((....(((((..............)))))))))))))))).(((...((.....))...))).......... ( -27.04) >DroSec_CAF1 58994 90 - 1 AUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAGGACCGAAGUGGGGACAUGGCCGGAAUG ........(((((((((((....(((((..............)))))))))))))))).......(((..(((....)))...))).... ( -34.24) >DroYak_CAF1 27597 90 - 1 AUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAGAACCGAAGUGGUAAUAUAGCAGAAAUG ........(((((((((((....(((((..............))))))))))))))))......(((.....)))............... ( -27.04) >consensus AUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAACAGGACCGAAGUGGGGAUAUGGCAGGAAUG ........(((((((((((....(((((..............)))))))))))))))).........(..(((....)))..)....... (-25.78 = -25.34 + -0.44)

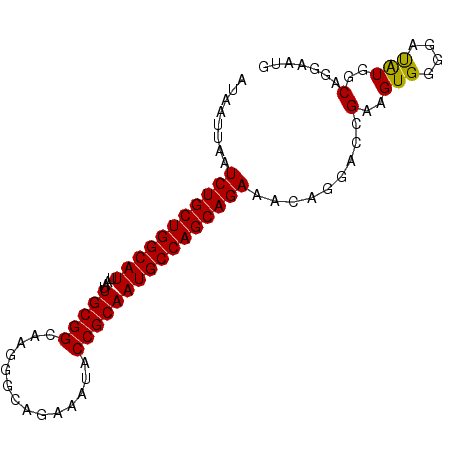
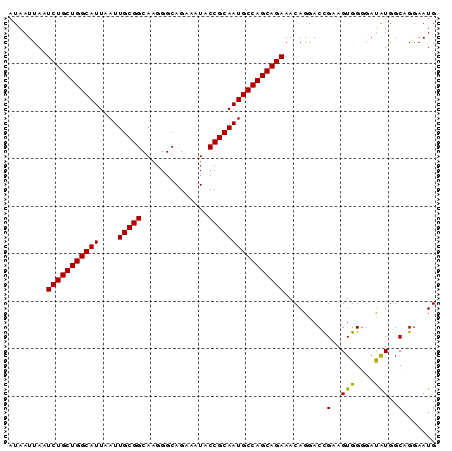
| Location | 1,340,611 – 1,340,710 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -38.10 |
| Energy contribution | -37.98 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340611 99 + 20766785 -------------------GUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGCUGCACACACUGCGUU--GAUGCGCGCAGCUU -------------------...((((((((((((((((((..........)))))))....)))))))))))..............((((((.......((((.--...)))))))))). ( -40.91) >DroSec_CAF1 59023 101 + 1 -------------------GUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGCUGCACACGCUGCGUGAAGCUGCGCGCAGCUU -------------------...((((((((((((((((((..........)))))))....)))))))))))..........(((....)))....((((((((......)))))))).. ( -46.00) >DroEre_CAF1 40459 117 + 1 AGUUUCUGUUUCCGUUUCCGUUUCUGCUGGCAUUGCGGUAUUUCUGCCUUUGCCGCAAUUAAUGCCAGUAGAUUAAUUAUACGCAUGGUUGCACACACUGUGUU--GGUGC-CGCAGCUU .....((((...(((.......((((((((((((((((((..........)))))))....)))))))))))........)))...(((.((((.....)))).--...))-)))))... ( -38.36) >DroYak_CAF1 27626 99 + 1 -------------------GUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGAUGCACACACUGCGUU--GAUGCGCGCAGCUU -------------------...((((((((((((((((((..........)))))))....))))))))))).........(((((.((((((.....))))))--.)))))........ ( -43.60) >consensus ___________________GUUUCUGCUGGCAUUGCGGUAUUUCUGCCCUUGCCGCAAUUAAUGCCAGCAGAUUAAUUAUACGCAUGGCUGCACACACUGCGUU__GAUGCGCGCAGCUU ......................((((((((((((((((((..........)))))))....)))))))))))..........(((....))).....((((((........))))))... (-38.10 = -37.98 + -0.12)

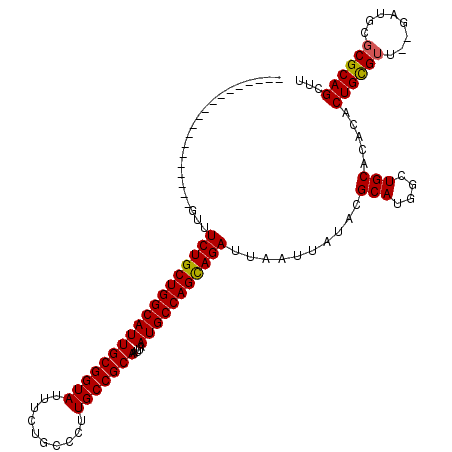
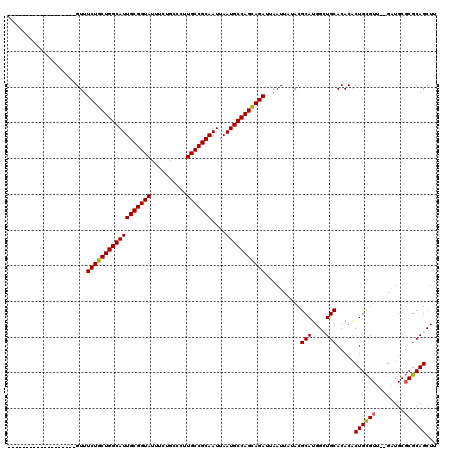
| Location | 1,340,611 – 1,340,710 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.15 |
| Mean single sequence MFE | -39.11 |
| Consensus MFE | -35.64 |
| Energy contribution | -37.02 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.13 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340611 99 - 20766785 AAGCUGCGCGCAUC--AACGCAGUGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAAC------------------- ..(((((((((((.--......)))))))))))...............(((((((((((....(((((..............))))))))))))))))...------------------- ( -43.14) >DroSec_CAF1 59023 101 - 1 AAGCUGCGCGCAGCUUCACGCAGCGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAAC------------------- ..(((((((((.(((......))))))))))))...............(((((((((((....(((((..............))))))))))))))))...------------------- ( -44.14) >DroEre_CAF1 40459 117 - 1 AAGCUGCG-GCACC--AACACAGUGUGUGCAACCAUGCGUAUAAUUAAUCUACUGGCAUUAAUUGCGGCAAAGGCAGAAAUACCGCAAUGCCAGCAGAAACGGAAACGGAAACAGAAACU ..((((..-(((((--(......)).))))...)).))((........(((.(((((((....(((((..............)))))))))))).)))..((....))...))....... ( -31.74) >DroYak_CAF1 27626 99 - 1 AAGCUGCGCGCAUC--AACGCAGUGUGUGCAUCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAAC------------------- ..(((((((((((.--......))))))))).....))..........(((((((((((....(((((..............))))))))))))))))...------------------- ( -37.44) >consensus AAGCUGCGCGCAUC__AACGCAGUGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAAUACCGCAAUGCCAGCAGAAAC___________________ ..(((((((((((.........)))))))))))...............(((((((((((....(((((..............))))))))))))))))...................... (-35.64 = -37.02 + 1.38)

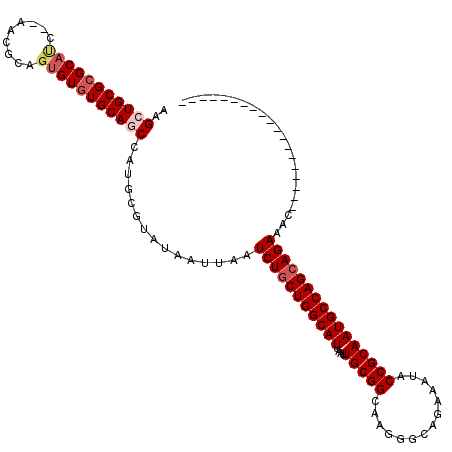
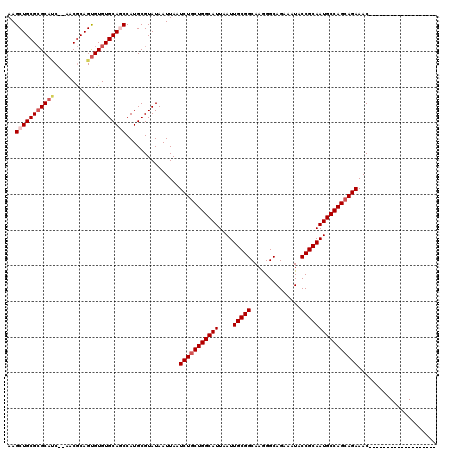
| Location | 1,340,632 – 1,340,750 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -23.83 |
| Energy contribution | -25.07 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340632 118 - 20766785 AUGUUUUUGGAAAUAAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCGCGCAUC--AACGCAGUGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAA .(((((((((((......)))))...))))))..((((....(((((((((((.--......))))))))))).................(((((.((.....))))))).))))..... ( -37.50) >DroSec_CAF1 59044 120 - 1 GUGCUUUUGGAAAUAAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCGCGCAGCUUCACGCAGCGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAA ((...(((((((......)))))))....))...((((.(((((((....))))))).((((((....)).((((.(((.((.....)).)))))))......))))....))))..... ( -38.80) >DroEre_CAF1 40499 109 - 1 -------U-UAAGUGAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCG-GCACC--AACACAGUGUGUGCAACCAUGCGUAUAAUUAAUCUACUGGCAUUAAUUGCGGCAAAGGCAGAAA -------.-.........................(((.....(((((.-(((((--(......)).))))............((((((((.....).))))))))))))...)))..... ( -20.00) >DroYak_CAF1 27647 110 - 1 -------U-UAAAUGAAAUUUUAAACAAAACAGAGCCUAGAAGCUGCGCGCAUC--AACGCAGUGUGUGCAUCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAA -------.-.........................((((....(((((((((((.--......))))))))).....))............(((((.((.....))))))).))))..... ( -25.10) >consensus _______U_GAAAUAAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCGCGCAUC__AACGCAGUGUGUGCAGCCAUGCGUAUAAUUAAUCUGCUGGCAUUAAUUGCGGCAAGGGCAGAAA ..................................((((....(((((((((((.........)))))))))))..(((...(((((((((.....).))))))))..))).))))..... (-23.83 = -25.07 + 1.25)

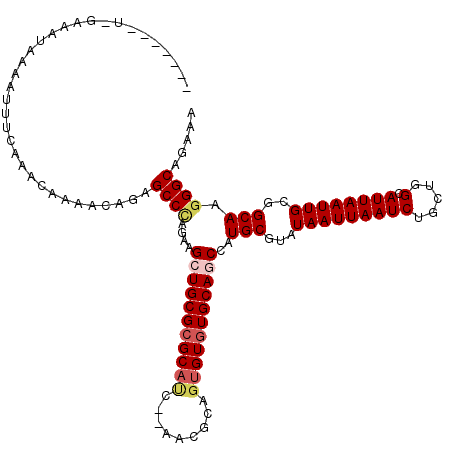
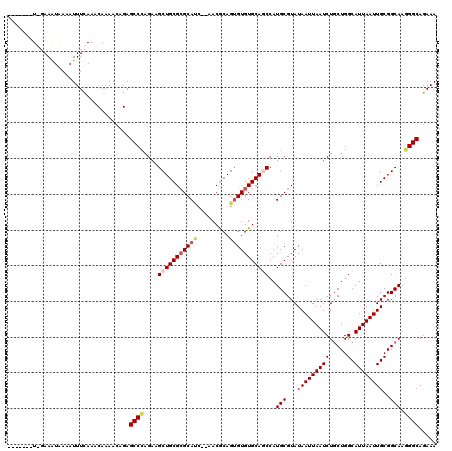
| Location | 1,340,672 – 1,340,790 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.89 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340672 118 + 20766785 ACGCAUGGCUGCACACACUGCGUU--GAUGCGCGCAGCUUCUGGGCUCUGUUUUGUUUGAAAUUUUAUUUCCAAAAACAUAAAACAAAAAACAAGUGAAAGGCACAAAGCCCAAAUACCA ......((((((.......((((.--...))))))))))..((((((.(((((((((((((((...)))))...)))))..)))))........(((.....)))..))))))....... ( -27.81) >DroSec_CAF1 59084 120 + 1 ACGCAUGGCUGCACACGCUGCGUGAAGCUGCGCGCAGCUUCUGGGCUCUGUUUUGUUUGAAAUUUUAUUUCCAAAAGCACAAAACAAAGAACAAGUGACAGACACAAAGCCCAAAUACCA (((((((........)).)))))(((((((....)))))))((((((.(((((((((((.((......)).)))....))))))))........(((.....)))..))))))....... ( -34.30) >DroEre_CAF1 40539 98 + 1 ACGCAUGGUUGCACACACUGUGUU--GGUGC-CGCAGCUUCUGGGCUCUGUUUUGUUUGAAAUUUCACUUA-A-------AAAACAA-----------ACGACACAAAGCCCAAAUACCA ......((((((...((((.....--)))).-.))))))..((((((.(((.(((((((............-.-------....)))-----------)))).))).))))))....... ( -28.03) >DroYak_CAF1 27687 110 + 1 ACGCAUGGAUGCACACACUGCGUU--GAUGCGCGCAGCUUCUAGGCUCUGUUUUGUUUAAAAUUUCAUUUA-A-------AAAACACAAAACAAUUGGAAGACACAAAGCCCAAAUACCA .(((((.((((((.....))))))--.)))))....((((((((....(((((((((((((......))))-)-------.....)))))))).))))))).)................. ( -26.70) >consensus ACGCAUGGCUGCACACACUGCGUU__GAUGCGCGCAGCUUCUGGGCUCUGUUUUGUUUGAAAUUUCAUUUA_A_______AAAACAAAAAACAAGUGAAAGACACAAAGCCCAAAUACCA ..(((....))).....((((((........))))))....((((((.((((((((((.......................)))))))...............))).))))))....... (-19.07 = -19.89 + 0.81)

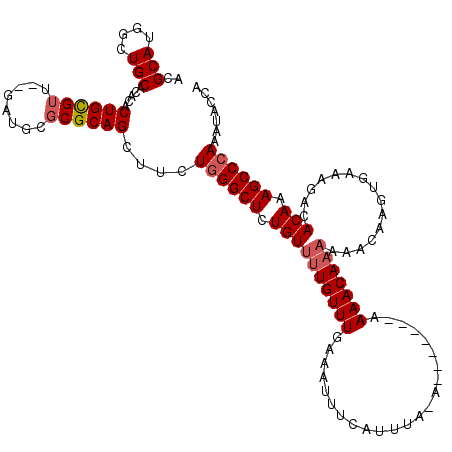
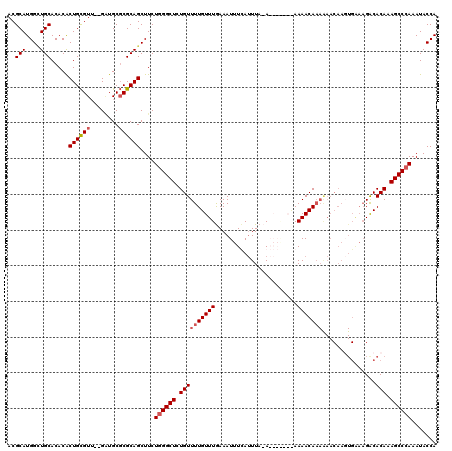
| Location | 1,340,672 – 1,340,790 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -29.05 |
| Energy contribution | -32.42 |
| Covariance contribution | 3.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340672 118 - 20766785 UGGUAUUUGGGCUUUGUGCCUUUCACUUGUUUUUUGUUUUAUGUUUUUGGAAAUAAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCGCGCAUC--AACGCAGUGUGUGCAGCCAUGCGU ..(((((((((((((((.........((((((...(((((((..........)))))))...)))))).)))))))))))).(((((((((((.--......)))))))))))..))).. ( -41.40) >DroSec_CAF1 59084 120 - 1 UGGUAUUUGGGCUUUGUGUCUGUCACUUGUUCUUUGUUUUGUGCUUUUGGAAAUAAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCGCGCAGCUUCACGCAGCGUGUGCAGCCAUGCGU ..((((((((((((((((.....))).........(((((((....((((((......))))))))))))))))))))))).(((((((((.(((......))))))))))))..))).. ( -43.40) >DroEre_CAF1 40539 98 - 1 UGGUAUUUGGGCUUUGUGUCGU-----------UUGUUUU-------U-UAAGUGAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCG-GCACC--AACACAGUGUGUGCAACCAUGCGU ((((.((((((((((((...((-----------(((..((-------(-(....))))...)))))...)))))))))))).......-(((((--(......)).)))).))))..... ( -33.30) >DroYak_CAF1 27687 110 - 1 UGGUAUUUGGGCUUUGUGUCUUCCAAUUGUUUUGUGUUUU-------U-UAAAUGAAAUUUUAAACAAAACAGAGCCUAGAAGCUGCGCGCAUC--AACGCAGUGUGUGCAUCCAUGCGU .(((.((((((((((((.........((((((...(((((-------.-.....)))))...)))))).)))))))))))).)))((((((((.--......)))))))).......... ( -33.10) >consensus UGGUAUUUGGGCUUUGUGUCUUUCACUUGUUUUUUGUUUU_______U_GAAAUAAAAUUUCAAACAAAACAGAGCCCAGAAGCUGCGCGCAUC__AACGCAGUGUGUGCAGCCAUGCGU ..(((((((((((((((.........((((((...(((((((..........)))))))...)))))).)))))))))))).(((((((((((.........)))))))))))..))).. (-29.05 = -32.42 + 3.38)

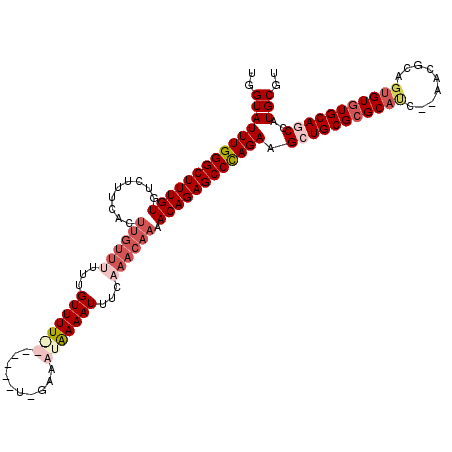
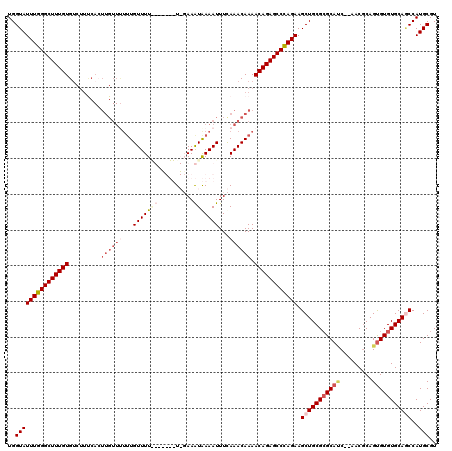
| Location | 1,340,710 – 1,340,830 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.61 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340710 120 + 20766785 CUGGGCUCUGUUUUGUUUGAAAUUUUAUUUCCAAAAACAUAAAACAAAAAACAAGUGAAAGGCACAAAGCCCAAAUACCAAACAGAAAAGAAGCUGCCUCCUAAUGAGUGCAAAUUAGCC ...((((((.(((((((((...((((((((......................))))))))(((.....))).......))))))))).)))...((((((.....))).))).....))) ( -23.55) >DroSec_CAF1 59124 120 + 1 CUGGGCUCUGUUUUGUUUGAAAUUUUAUUUCCAAAAGCACAAAACAAAGAACAAGUGACAGACACAAAGCCCAAAUACCAGCCAGAAGAGCAGCUGCCUCCUAAUGAGUGCAAAUUAGCC .((((((.(((((((((((.((......)).)))....))))))))........(((.....)))..)))))).......(((....).))...((((((.....))).)))........ ( -23.00) >DroEre_CAF1 40576 100 + 1 CUGGGCUCUGUUUUGUUUGAAAUUUCACUUA-A-------AAAACAA-----------ACGACACAAAGCCCAAAUACCAAGCAAA-AAGUAGCUGCCUCCUAAUGAGUGCAAAUUAGCC .((((((.(((.(((((((............-.-------....)))-----------)))).))).)))))).......(((...-.....)))(((((.....))).))......... ( -24.53) >DroYak_CAF1 27725 112 + 1 CUAGGCUCUGUUUUGUUUAAAAUUUCAUUUA-A-------AAAACACAAAACAAUUGGAAGACACAAAGCCCAAAUACCAACCAAAAAAGGACCUGCCUCCUAAUGAGUGCAAAUUAGCC ...((((.(((((((((((((......))))-)-------.....)))))))).(((.......)))))))..........((......))...((((((.....))).)))........ ( -16.90) >consensus CUGGGCUCUGUUUUGUUUGAAAUUUCAUUUA_A_______AAAACAAAAAACAAGUGAAAGACACAAAGCCCAAAUACCAACCAAAAAAGAAGCUGCCUCCUAAUGAGUGCAAAUUAGCC .((((((.((((((((((.......................)))))))...............))).)))))).....................((((((.....))).)))........ (-14.86 = -15.61 + 0.75)

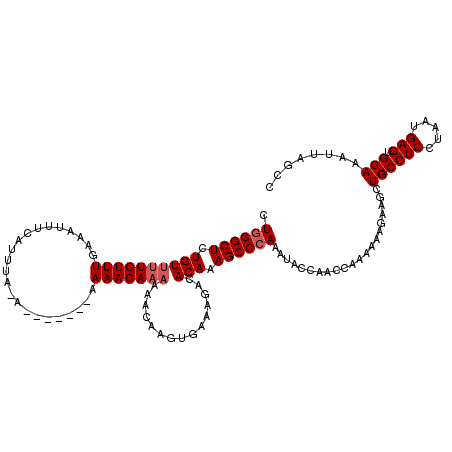
| Location | 1,340,710 – 1,340,830 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -20.70 |
| Energy contribution | -23.70 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340710 120 - 20766785 GGCUAAUUUGCACUCAUUAGGAGGCAGCUUCUUUUCUGUUUGGUAUUUGGGCUUUGUGCCUUUCACUUGUUUUUUGUUUUAUGUUUUUGGAAAUAAAAUUUCAAACAAAACAGAGCCCAG (((....((((.(((.....))))))).......((((((((((....((((.....))))...)))(((((...(((((((..........)))))))...)))))))))))))))... ( -28.60) >DroSec_CAF1 59124 120 - 1 GGCUAAUUUGCACUCAUUAGGAGGCAGCUGCUCUUCUGGCUGGUAUUUGGGCUUUGUGUCUGUCACUUGUUCUUUGUUUUGUGCUUUUGGAAAUAAAAUUUCAAACAAAACAGAGCCCAG .((......))(((..(((((((((....)).)))))))..)))...(((((((((((.....))).........(((((((....((((((......))))))))))))))))))))). ( -33.70) >DroEre_CAF1 40576 100 - 1 GGCUAAUUUGCACUCAUUAGGAGGCAGCUACUU-UUUGCUUGGUAUUUGGGCUUUGUGUCGU-----------UUGUUUU-------U-UAAGUGAAAUUUCAAACAAAACAGAGCCCAG .(((((..(((.(((.....))))))((.....-...)))))))...((((((((((...((-----------(((..((-------(-(....))))...)))))...)))))))))). ( -31.50) >DroYak_CAF1 27725 112 - 1 GGCUAAUUUGCACUCAUUAGGAGGCAGGUCCUUUUUUGGUUGGUAUUUGGGCUUUGUGUCUUCCAAUUGUUUUGUGUUUU-------U-UAAAUGAAAUUUUAAACAAAACAGAGCCUAG .((((((((((.(((.....)))))))))((......)).))))...((((((((((.........((((((...(((((-------.-.....)))))...)))))).)))))))))). ( -28.10) >consensus GGCUAAUUUGCACUCAUUAGGAGGCAGCUACUUUUCUGGUUGGUAUUUGGGCUUUGUGUCUUUCACUUGUUUUUUGUUUU_______U_GAAAUAAAAUUUCAAACAAAACAGAGCCCAG .((((((((((.(((.....)))))))...........))))))...((((((((((.........((((((...(((((((..........)))))))...)))))).)))))))))). (-20.70 = -23.70 + 3.00)

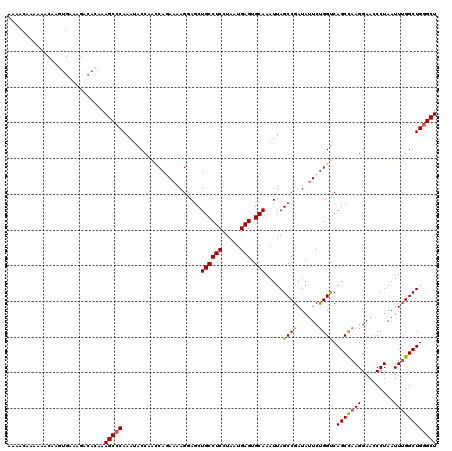
| Location | 1,340,750 – 1,340,870 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.26 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340750 120 + 20766785 AAAACAAAAAACAAGUGAAAGGCACAAAGCCCAAAUACCAAACAGAAAAGAAGCUGCCUCCUAAUGAGUGCAAAUUAGCCGAUAUUCUGGUCAGCUAAGGAACCCUAAUUUGGCUGGGCU ..............(((.....)))..((((((....(((((............((((((.....))).)))..(((((.(((......))).)))))..........))))).)))))) ( -26.80) >DroSec_CAF1 59164 120 + 1 AAAACAAAGAACAAGUGACAGACACAAAGCCCAAAUACCAGCCAGAAGAGCAGCUGCCUCCUAAUGAGUGCAAAUUAGCCGAUAUUCUGGUCAGCCAAGGAAACCUAAUUUGGCUGUGCU ..............(((.....)))..(((..........(((((((...(.((((((((.....))).)).....))).)...)))))))((((((((....).....))))))).))) ( -29.00) >DroSim_CAF1 37281 118 + 1 GGA-C-AAAAACAAGUGACAGACACAAAGCCCAAAUACCAGCCAGAAGAGGAGCUGCCUCCUAAUGAGUGCAAAUUAGCCGAUAUUCUGGUCAGCCAAGGAACCCUAAUUUGGCUGGGCU ...-.-........(((.....)))..(((((........(((((((..((...((((((.....))).)))......))....))))))).(((((((.........)))))))))))) ( -34.30) >DroEre_CAF1 40608 108 + 1 AAAACAA-----------ACGACACAAAGCCCAAAUACCAAGCAAA-AAGUAGCUGCCUCCUAAUGAGUGCAAAUUAGCCGAUACUCCGGCCAGCCAAGGAACCCUAAUUGGGCUGGGCU .......-----------.........((((((....((..((...-.......((((((.....))).))).....((((......))))..))...))..(((.....))).)))))) ( -28.30) >DroYak_CAF1 27757 120 + 1 AAAACACAAAACAAUUGGAAGACACAAAGCCCAAAUACCAACCAAAAAAGGACCUGCCUCCUAAUGAGUGCAAAUUAGCCGAUAUUCUGGUCAGCUAAGGAACCCUAAUUUGGCUGGGCU ..............(((.......)))((((((........(((((..(((.((((((((.....))).)))..(((((.(((......))).)))))))...)))..))))).)))))) ( -27.20) >consensus AAAACAAAAAACAAGUGAAAGACACAAAGCCCAAAUACCAACCAGAAAAGGAGCUGCCUCCUAAUGAGUGCAAAUUAGCCGAUAUUCUGGUCAGCCAAGGAACCCUAAUUUGGCUGGGCU ...........................(((((......................((((((.....))).))).....((((......)))).(((((((.........)))))))))))) (-21.42 = -21.26 + -0.16)

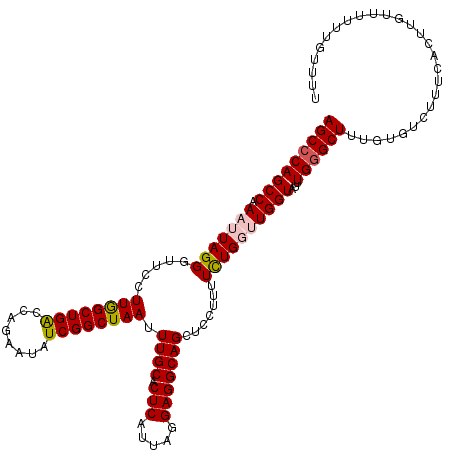
| Location | 1,340,750 – 1,340,870 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1340750 120 - 20766785 AGCCCAGCCAAAUUAGGGUUCCUUAGCUGACCAGAAUAUCGGCUAAUUUGCACUCAUUAGGAGGCAGCUUCUUUUCUGUUUGGUAUUUGGGCUUUGUGCCUUUCACUUGUUUUUUGUUUU (((((((((((((.((((....((((((((........)))))))).((((.(((.....)))))))......)))))))))))...))))))..(((.....))).............. ( -32.50) >DroSec_CAF1 59164 120 - 1 AGCACAGCCAAAUUAGGUUUCCUUGGCUGACCAGAAUAUCGGCUAAUUUGCACUCAUUAGGAGGCAGCUGCUCUUCUGGCUGGUAUUUGGGCUUUGUGUCUGUCACUUGUUCUUUGUUUU (((.((((((....(((...))))))))).(((((.(((((((((..((((.(((.....))))))).........)))))))))))))))))..(((.....))).............. ( -32.80) >DroSim_CAF1 37281 118 - 1 AGCCCAGCCAAAUUAGGGUUCCUUGGCUGACCAGAAUAUCGGCUAAUUUGCACUCAUUAGGAGGCAGCUCCUCUUCUGGCUGGUAUUUGGGCUUUGUGUCUGUCACUUGUUUUU-G-UCC ...(((((((.(..((((....((((((((........)))))))).((((.(((.....))))))).))))..).))))))).....((((...(((.....)))........-)-))) ( -34.50) >DroEre_CAF1 40608 108 - 1 AGCCCAGCCCAAUUAGGGUUCCUUGGCUGGCCGGAGUAUCGGCUAAUUUGCACUCAUUAGGAGGCAGCUACUU-UUUGCUUGGUAUUUGGGCUUUGUGUCGU-----------UUGUUUU ((((((((((.....)))).((..(((((((((......))))))..((((.(((.....)))))))......-...))).))....)))))).........-----------....... ( -33.20) >DroYak_CAF1 27757 120 - 1 AGCCCAGCCAAAUUAGGGUUCCUUAGCUGACCAGAAUAUCGGCUAAUUUGCACUCAUUAGGAGGCAGGUCCUUUUUUGGUUGGUAUUUGGGCUUUGUGUCUUCCAAUUGUUUUGUGUUUU ...(((((((((..((((..((((((((((........))))))))..(((.(((.....))))))))))))..))))))))).....((((.....))))................... ( -35.90) >consensus AGCCCAGCCAAAUUAGGGUUCCUUGGCUGACCAGAAUAUCGGCUAAUUUGCACUCAUUAGGAGGCAGCUCCUUUUCUGGUUGGUAUUUGGGCUUUGUGUCUUUCACUUGUUUUUUGUUUU (((((((((.(((((((.....((((((((........)))))))).((((.(((.....))))))).......))))))))))...))))))........................... (-26.66 = -27.02 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:39 2006