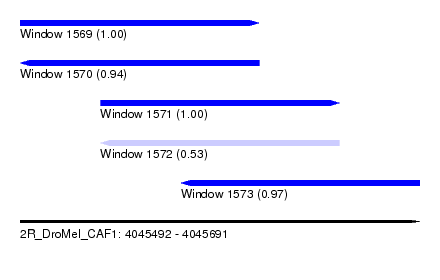
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,045,492 – 4,045,691 |
| Length | 199 |
| Max. P | 0.997940 |

| Location | 4,045,492 – 4,045,611 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -27.64 |
| Energy contribution | -28.44 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4045492 119 + 20766785 GCUGCAAAUGAAGUGCAAAAAAAUUGUUAAACGCUUUACGCACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA-A (((.(....)..(((((((....(((((...((((((((((....((.(((.....))).))....)))))))))).((((......))))..)))))...)))))))...)))....-. ( -30.40) >DroSec_CAF1 28139 118 + 1 GCUGCAAAUGAAGUGCAAA-AAAUUGUUAAACGCUUUACGCACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA-A (((.(....)..(((((((-.(.(((((...((((((((((....((.(((.....))).))....)))))))))).((((......))))..))))).).)))))))...)))....-. ( -31.10) >DroSim_CAF1 32807 118 + 1 GCUGCAAAUGAAGUGCAAA-AAAUUGUUAAACGCUUUACGCACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA-A (((.(....)..(((((((-.(.(((((...((((((((((....((.(((.....))).))....)))))))))).((((......))))..))))).).)))))))...)))....-. ( -31.10) >DroEre_CAF1 27433 118 + 1 GCUGCAAAUGAAGUGCAAC-AAAUUGUUAAACGCUUUACGCACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUGGA-A (((.(....)..((((((.-...(((((...((((((((((....((.(((.....))).))....)))))))))).((((......))))..)))))....))))))...)))....-. ( -29.90) >DroYak_CAF1 31330 119 + 1 GCUGCAAAUGAAGUGCAAC-AAAUUGUUAAACGCUUAACGCACAAUUAGGCAGUCAGCCAUAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGAACGAGCGCUUGAAA ........(.(((((((((-.....)))....(((((((((((..(((.((.(((........))))).))).)))))).....))))).....................)))))).).. ( -26.50) >consensus GCUGCAAAUGAAGUGCAAA_AAAUUGUUAAACGCUUUACGCACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA_A ............(((((((....(((((...((((((((((.......(((.....))).......)))))))))).((((......))))..)))))...)))))))............ (-27.64 = -28.44 + 0.80)

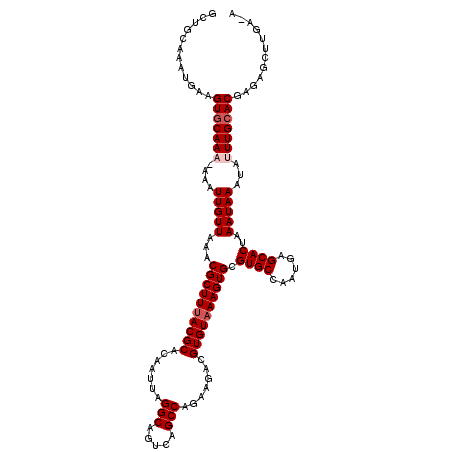
| Location | 4,045,492 – 4,045,611 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4045492 119 - 20766785 U-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUGCGUAAAGCGUUUAACAAUUUUUUUGCACUUCAUUUGCAGC .-....((....(((((((..........(((((....)))))((.((((((.((......(((.....)))......)).)))))).))...........))))))).......))... ( -27.70) >DroSec_CAF1 28139 118 - 1 U-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUGCGUAAAGCGUUUAACAAUUU-UUUGCACUUCAUUUGCAGC .-....((....(((((((..........(((((....)))))((.((((((.((......(((.....)))......)).)))))).))..........-))))))).......))... ( -27.70) >DroSim_CAF1 32807 118 - 1 U-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUGCGUAAAGCGUUUAACAAUUU-UUUGCACUUCAUUUGCAGC .-....((....(((((((..........(((((....)))))((.((((((.((......(((.....)))......)).)))))).))..........-))))))).......))... ( -27.70) >DroEre_CAF1 27433 118 - 1 U-UCCAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUGCGUAAAGCGUUUAACAAUUU-GUUGCACUUCAUUUGCAGC .-....((....((((((...........(((((....)))))((.((((((.((......(((.....)))......)).)))))).))..........-.)))))).......))... ( -27.00) >DroYak_CAF1 31330 119 - 1 UUUCAAGCGCUCGUUCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUAUGGCUGACUGCCUAAUUGUGCGUUAAGCGUUUAACAAUUU-GUUGCACUUCAUUUGCAGC ....((((((((((.(((...........(((((....)))))..................(((.....)))...))).)))...)))))))........-((((((.......)))))) ( -26.90) >consensus U_UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUGCGUAAAGCGUUUAACAAUUU_UUUGCACUUCAUUUGCAGC ......((....((((((...........(((((....)))))((.((((((.((......(((.....)))......)).)))))).))............)))))).......))... (-24.92 = -25.32 + 0.40)

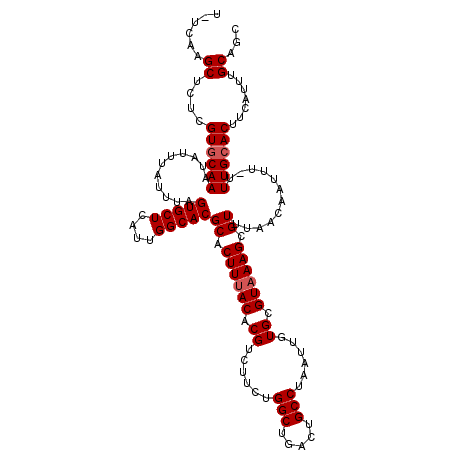
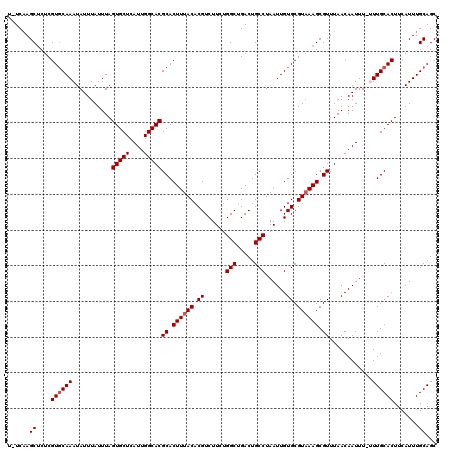
| Location | 4,045,532 – 4,045,651 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4045532 119 + 20766785 CACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA-AAAAACAGCCCGAGUCCAAGAACAAGGACAAUCCGGGUAAG .....((((((.(((........)))(((((((((((..........)))))..........))))))....))))))-.......(((((.((((........))))....)))))... ( -31.50) >DroSec_CAF1 28178 119 + 1 CACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA-AAAAACAGCCCGAGUCCAAGAACAAGGACAAUCCGGGUAAG .....((((((.(((........)))(((((((((((..........)))))..........))))))....))))))-.......(((((.((((........))))....)))))... ( -31.50) >DroSim_CAF1 32846 119 + 1 CACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA-AAAAACAGCCCGAGUCCAAGAACAAGGACAAUCCGGGUAAG .....((((((.(((........)))(((((((((((..........)))))..........))))))....))))))-.......(((((.((((........))))....)))))... ( -31.50) >DroEre_CAF1 27472 119 + 1 CACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUGGA-AAAAGCAGCCCGAGUCCAAGAACAAGGACAAUCCGGGUAAG ........(((.....)))......((((((((((((..........)))))..........)))))))...(((...-...))).(((((.((((........))))....)))))... ( -32.70) >DroYak_CAF1 31369 120 + 1 CACAAUUAGGCAGUCAGCCAUAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGAACGAGCGCUUGAAAAAAACAGCUCGAGUCCAAGAACAAGGACAAACCGGGUAAG (((..((((((.....))).)))...)))...(((((..........))))).......((((((..(((((..((....))....))))).((((........))))....)))))).. ( -27.80) >consensus CACAAUUAGGCAGUCAGCCAGAAGACGUGUAAAGUGCGUGCCAAUGAGCACUAAAUAAAUAUUUGCACGAGAGCUUGA_AAAAACAGCCCGAGUCCAAGAACAAGGACAAUCCGGGUAAG .....((((((.(((........)))(((((((((((..........)))))..........))))))....))))))........(((((.((((........))))....)))))... (-28.80 = -29.04 + 0.24)

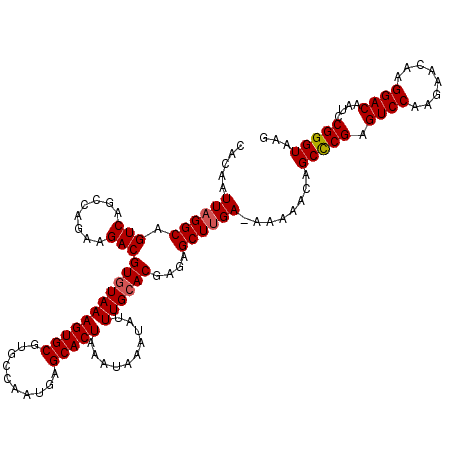
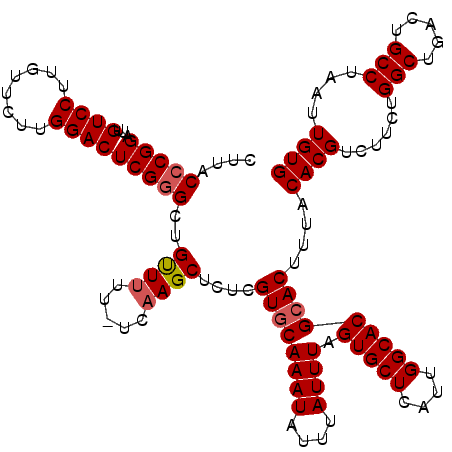
| Location | 4,045,532 – 4,045,651 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.16 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4045532 119 - 20766785 CUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUG ....(((((...((((........)))))))))........-..........((((((((....)))).(((((....)))))))))....((((......(((.....)))....)))) ( -29.50) >DroSec_CAF1 28178 119 - 1 CUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUG ....(((((...((((........)))))))))........-..........((((((((....)))).(((((....)))))))))....((((......(((.....)))....)))) ( -29.50) >DroSim_CAF1 32846 119 - 1 CUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUG ....(((((...((((........)))))))))........-..........((((((((....)))).(((((....)))))))))....((((......(((.....)))....)))) ( -29.50) >DroEre_CAF1 27472 119 - 1 CUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGCUUUU-UCCAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUG ....(((((...((((........)))))))))..(((...-...)))....((((((((....)))).(((((....)))))))))....((((......(((.....)))....)))) ( -30.90) >DroYak_CAF1 31369 120 - 1 CUUACCCGGUUUGUCCUUGUUCUUGGACUCGAGCUGUUUUUUUCAAGCGCUCGUUCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUAUGGCUGACUGCCUAAUUGUG ............((((........)))).(((((.((((.....)))))))))................(((((....)))))........((((..(((.(((.....)))))).)))) ( -26.30) >consensus CUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU_UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGGCACGCACUUUACACGUCUUCUGGCUGACUGCCUAAUUGUG ....(((((...((((........)))))))))..((((.....))))....((((((((....)))).(((((....)))))))))....((((......(((.....)))....)))) (-26.72 = -27.16 + 0.44)

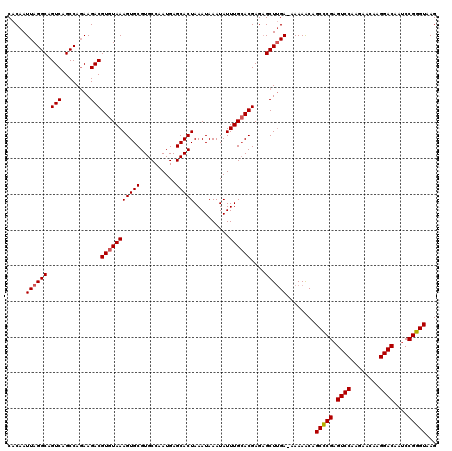
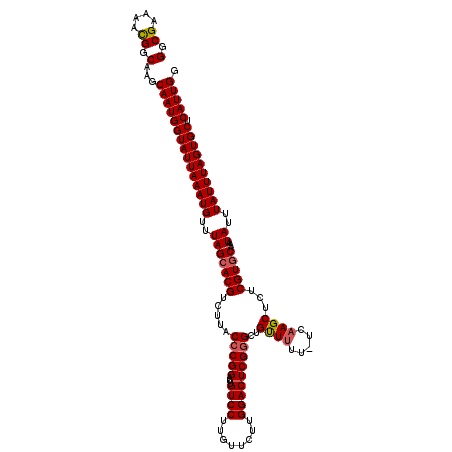
| Location | 4,045,572 – 4,045,691 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -31.72 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4045572 119 - 20766785 GGCGAAAAUGGCAAGCAAUGGUAUUAAAUGUUUAGCACGUCUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGG .((.......))...(((((((((((((((..(((((((.....(((((...((((........)))))))))..((((..-..))))...)))))...))..)))))))))).))))). ( -33.60) >DroSec_CAF1 28218 119 - 1 GGCGAAAACGGCAAGCAAUGGUAUUAAAUGUUUAGCACGUCUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGG (.((....)).)...(((((((((((((((..(((((((.....(((((...((((........)))))))))..((((..-..))))...)))))...))..)))))))))).))))). ( -34.50) >DroSim_CAF1 32886 119 - 1 GGCGAAAACGGCAAGCAAUGGUAUUAAAUGUUUAGCACGUCUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU-UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGG (.((....)).)...(((((((((((((((..(((((((.....(((((...((((........)))))))))..((((..-..))))...)))))...))..)))))))))).))))). ( -34.50) >DroEre_CAF1 27512 119 - 1 GGCGAAAACGGCAAGCAAUGGUAUUAAAUGUUUAGCACGUCUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGCUUUU-UCCAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGG (.((....)).)...(((((((((((((((..(((((((.....(((((...((((........)))))))))..(((...-...)))...)))))...))..)))))))))).))))). ( -36.20) >DroYak_CAF1 31409 120 - 1 GGCGAAAACGGCAAGCAAUGGUAUUAAAUGUUUAGUACGUCUUACCCGGUUUGUCCUUGUUCUUGGACUCGAGCUGUUUUUUUCAAGCGCUCGUUCAAAUAUUUAUUUAGUGCUCAUUGG (.((....)).)...(((((((((((((((..((..................((((........)))).(((((.((((.....)))))))))......))..)))))))))).))))). ( -29.90) >consensus GGCGAAAACGGCAAGCAAUGGUAUUAAAUGUUUAGCACGUCUUACCCGGAUUGUCCUUGUUCUUGGACUCGGGCUGUUUUU_UCAAGCUCUCGUGCAAAUAUUUAUUUAGUGCUCAUUGG (.((....)).)...(((((((((((((((..(((((((.....(((((...((((........)))))))))..((((.....))))...)))))...))..)))))))))).))))). (-31.72 = -32.00 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:32 2006