
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,043,340 – 4,043,539 |
| Length | 199 |
| Max. P | 0.999821 |

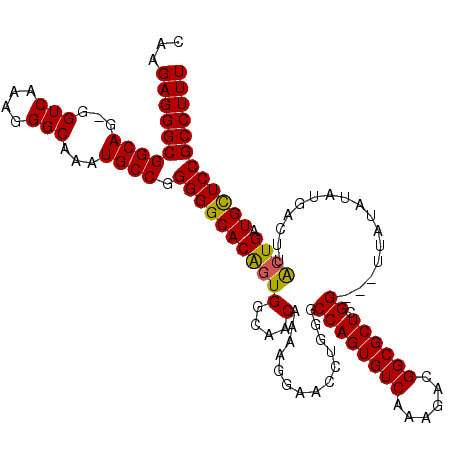
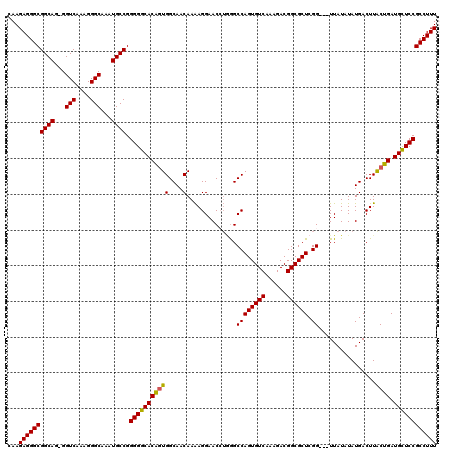
| Location | 4,043,340 – 4,043,459 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -45.48 |
| Consensus MFE | -40.30 |
| Energy contribution | -40.02 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4043340 119 - 20766785 CAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUGGGCCAGUGUCAAAGACGGCGCUCGGCGCUUAUAUAUGACUUACUGAUGCUCCGCCUUU ...((((((((.((-.((((..(((((.((((((((..(....((....))....)..)))))(((((((((......)))))).))).....))).)).)))..)))).)))))))))) ( -48.50) >DroSec_CAF1 26013 116 - 1 CAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUUGGCCAGUGUCAAAGACGGCGCUCGG---UUAUAUAUGACUUACUGAUGCUCCGCCUUU ...((((((((((.-.(((....)))...)))).(((((((((((....)..(((......(((((((((((......)))))).))---))).......))))))).)))))))))))) ( -46.42) >DroSim_CAF1 28175 116 - 1 CAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUGGGCCAGUGUCAAAGACGGCGCUCGG---UUAUAUAUGACUUACUGAUGCUCCGCCUUU ...((((((((.((-.((((..(((((.((((((((..(....((....))....)..)))))(((((((((......)))))).))---)..))).)).)))..)))).)))))))))) ( -45.90) >DroEre_CAF1 25142 116 - 1 CAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACGUUGGCAACAAGAGGAACGGGAGCCAGUGUCAAAGACGGCGCUCGG---UUAUAUAUGACUUACUGAUGCUCCGCCUUU ...((((((((((.-.(((....)))...)))).((((((..(((....)))(((.......((((((((((......)))))).))---))........))).....)))))))))))) ( -43.06) >DroYak_CAF1 29082 117 - 1 AAAGAGGGCGGCAGGGGUCAAAGGGCAAGUGCCGGGGGCACAGUGGCAACAAAAGGAACGUAGGCCAGUGUCAAAGACGGCGCUCGG---UUAUAUAUGACUUGCUGAUGUUCCGCCUUU ...((((((((((...(((....)))...)))).(((((((((((....)..(((..(..((((((((((((......)))))).))---)).))..)..))))))).)))))))))))) ( -43.50) >consensus CAAGAGGGCGGCAG_GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUGGGCCAGUGUCAAAGACGGCGCUCGG___UUAUAUAUGACUUACUGAUGCUCCGCCUUU ...((((((((((...(((....)))...)))).(((((((((((....)..............((((((((......)))))).))................)))).)))))))))))) (-40.30 = -40.02 + -0.28)

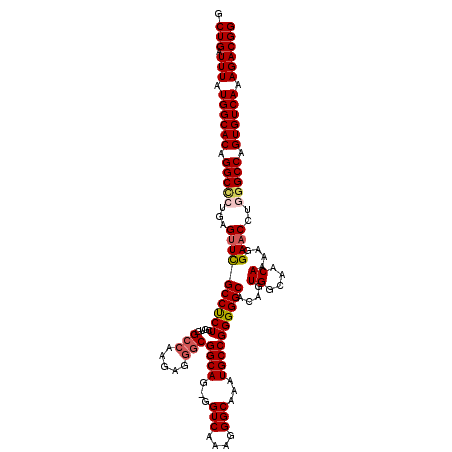
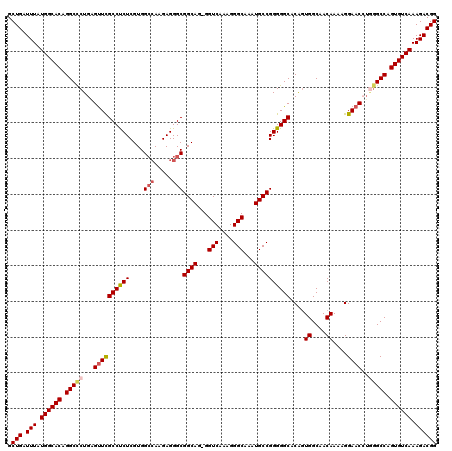
| Location | 4,043,380 – 4,043,499 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -50.50 |
| Consensus MFE | -45.44 |
| Energy contribution | -46.20 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4043380 119 - 20766785 GCUGAUUUAUGGCACGGGCCCUGAGUUCGCCUCUCGUGGCCAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUGGGCCAGUGUCAAAGACGG .(((.(((.((((((.(((((.(.((((((((((....(((.....)))((((.-.(((....)))...))))))))))....((....))....))))).))))).)))))).)))))) ( -52.90) >DroSec_CAF1 26050 119 - 1 GCUGAUUUAUGGCACAGGCCCUGAGUUCGCCUCUCGUGGCCAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUUGGCCAGUGUCAAAGACGG .(((.(((.((((((.((((..(.((((((((((....(((.....)))((((.-.(((....)))...))))))))))....((....))....)))))..)))).)))))).)))))) ( -48.80) >DroSim_CAF1 28212 119 - 1 GCUGAUUUAUGGCACAGGCCCUGAGUUCGCCUCUCGUGGCCAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUGGGCCAGUGUCAAAGACGG .(((.(((.((((((.(((((.(.((((((((((....(((.....)))((((.-.(((....)))...))))))))))....((....))....))))).))))).)))))).)))))) ( -52.20) >DroEre_CAF1 25179 118 - 1 GCUGAUUUAUGGCACAGGCCCUG-GUUCGCCCCUCGUGGCCAAGAGGGCGGCAG-GGUCAAAGGGCAAAUGCCGGGGGCACGUUGGCAACAAGAGGAACGGGAGCCAGUGUCAAAGACGG .(((.(((.((((((.(((.((.-((((((((((....(((.....)))((((.-.(((....)))...))))))))))...(((....)))...)))).)).))).)))))).)))))) ( -53.20) >DroYak_CAF1 29119 120 - 1 GCUGAUUUAUGGCACAGGCUCUGGGCUUGCCUCUCGUGGAAAAGAGGGCGGCAGGGGUCAAAGGGCAAGUGCCGGGGGCACAGUGGCAACAAAAGGAACGUAGGCCAGUGUCAAAGACGG .(((.(((.((((((.((((((..(((.(((.(((........))))))))).))))))....(((..(((((...)))))..((....))............))).)))))).)))))) ( -45.40) >consensus GCUGAUUUAUGGCACAGGCCCUGAGUUCGCCUCUCGUGGCCAAGAGGGCGGCAG_GGUCAAAGGGCAAAUGCCGGGGGCACAGUGGCAACAAAAGGAACCUGGGCCAGUGUCAAAGACGG .(((.(((.((((((.(((((...((((((((((....(((.....)))((((...(((....)))...))))))))))....((....))....))))..))))).)))))).)))))) (-45.44 = -46.20 + 0.76)

| Location | 4,043,420 – 4,043,539 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.06 |
| Mean single sequence MFE | -38.39 |
| Consensus MFE | -35.96 |
| Energy contribution | -35.88 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4043420 119 + 20766785 UGCCCCCGGCAUUUGCCCUUUGACC-CUGCCGCCCUCUUGGCCACGAGAGGCGAACUCAGGGCCCGUGCCAUAAAUCAGCUUUGUUGCAUUUAAGUCGGUGCAAACAAGCUCACAACAAU .......(((((..(((((..((..-....((((.(((((....)))))))))...)))))))..))))).......(((((..(((((((......)))))))..)))))......... ( -39.10) >DroSec_CAF1 26090 119 + 1 UGCCCCCGGCAUUUGCCCUUUGACC-CUGCCGCCCUCUUGGCCACGAGAGGCGAACUCAGGGCCUGUGCCAUAAAUCAGCUUUGUUGCAUUUAAGUCGGUGCAAACAAGCUCACAACAAU .......(((((..(((((..((..-....((((.(((((....)))))))))...)))))))..))))).......(((((..(((((((......)))))))..)))))......... ( -40.00) >DroSim_CAF1 28252 119 + 1 UGCCCCCGGCAUUUGCCCUUUGACC-CUGCCGCCCUCUUGGCCACGAGAGGCGAACUCAGGGCCUGUGCCAUAAAUCAGCUUUGUUGCAUUUAAGUCGGUGCAAACAAGCUCACAACAAU .......(((((..(((((..((..-....((((.(((((....)))))))))...)))))))..))))).......(((((..(((((((......)))))))..)))))......... ( -40.00) >DroEre_CAF1 25219 118 + 1 UGCCCCCGGCAUUUGCCCUUUGACC-CUGCCGCCCUCUUGGCCACGAGGGGCGAAC-CAGGGCCUGUGCCAUAAAUCAGCUUUGUUGCAUUUAAGUCGGUGCAAACAAGCUCACAACAAU .......(((((..(((((......-....(((((.((((....)))))))))...-.)))))..))))).......(((((..(((((((......)))))))..)))))......... ( -41.04) >DroYak_CAF1 29159 120 + 1 UGCCCCCGGCACUUGCCCUUUGACCCCUGCCGCCCUCUUUUCCACGAGAGGCAAGCCCAGAGCCUGUGCCAUAAAUCAGCUUUGUUGCAUUUAAGUCAGUGCAAACAAGCUCACAACAAU .......(((((..((..((((..(..((((...(((........))).)))).)..))))))..))))).......(((((..(((((((......)))))))..)))))......... ( -31.80) >consensus UGCCCCCGGCAUUUGCCCUUUGACC_CUGCCGCCCUCUUGGCCACGAGAGGCGAACUCAGGGCCUGUGCCAUAAAUCAGCUUUGUUGCAUUUAAGUCGGUGCAAACAAGCUCACAACAAU .......(((((..(((((..(........)(((.(((((....))))))))......)))))..))))).......(((((..(((((((......)))))))..)))))......... (-35.96 = -35.88 + -0.08)

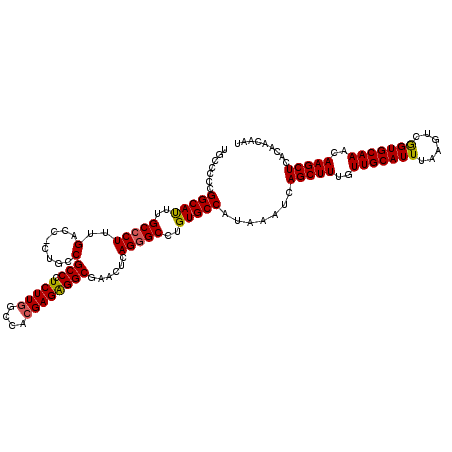
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:26 2006