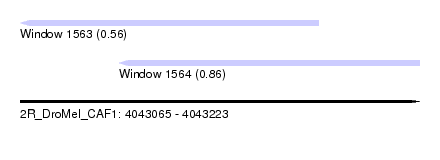
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,043,065 – 4,043,223 |
| Length | 158 |
| Max. P | 0.864990 |

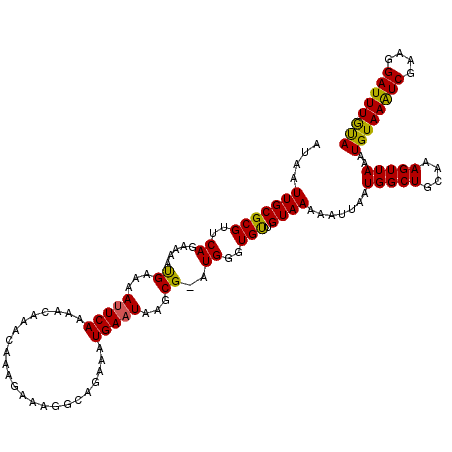
| Location | 4,043,065 – 4,043,183 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -17.66 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4043065 118 - 20766785 AGAAAGGCAGAAAUGAAUAAGCG-AUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGUAUAAUAUAAAUUGGCAA-AUUUCCCGAAACGAACUGAAGCG .((((.(((....).......((-((..((((((((........(((((....)))))....((((((....))))))))))))))..))))))..-.)))).................. ( -19.00) >DroSec_CAF1 25740 118 - 1 AGAAAGGCAGA-AUGAAUAAGCGGAUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGUAUAAUAAAAAUUGGCAA-AUUUCCCGAAACGAGCCGAAGCG .....(((...-........((.(((...(((((((........(((((....)))))....((((((....)))))))))))))...))).))..-......((...)).)))...... ( -22.00) >DroSim_CAF1 27901 120 - 1 AGAAAGGCAGAAAUGAAUAAGCGGAUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUAUAUAAUAUAAAUUGGCAAAAUUUCCCGAAACGAGCCGAAGCG .....(((.(((((......((.(((..((((((((........(((((....)))))....((((((....))))))))))))))..))).))...))))).((...)).)))...... ( -25.20) >DroEre_CAF1 24885 118 - 1 AGAAAGGCGGGAAUGAAUAAGCG-AUGGGUGCUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGCAUAAUAUAAAUUGCCAA-AUUUCCCGAAACGAGCUGAAGCG .....(.((((((.......(((-((..(((.............(((((....))))).(((((((((....)))))))))..)))..)))))...-..))))))...)..((....)). ( -29.50) >DroYak_CAF1 28821 118 - 1 AGAAUGGAAGGAAUGAAUAAGCG-AUGGGUGUUGUAAAAAUUAAUGGCUGAAAAGUUAAAUGUAAGUCGUAGGAUUUGCAUAAUAUAAACUGUCAC-AUUUCUCGAAACGAGCUGAAGCG ....................(((-((((...(((((........(((((....))))).(((((((((....)))))))))..))))).)))))..-....((((...)))).....)). ( -23.80) >consensus AGAAAGGCAGAAAUGAAUAAGCG_AUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGUAUAAUAUAAAUUGGCAA_AUUUCCCGAAACGAGCUGAAGCG ....................((...(.(((.((((.........(((((....))))).(((((((((....)))))))))..........................))))))).).)). (-17.66 = -16.90 + -0.76)

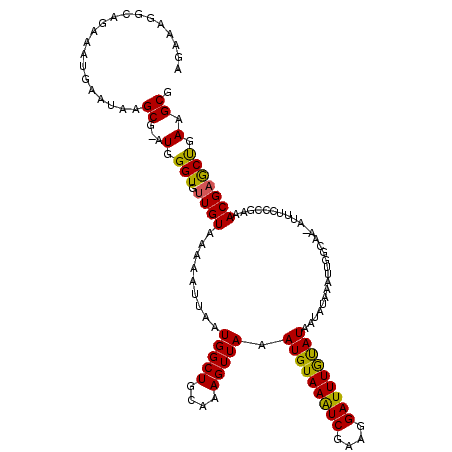
| Location | 4,043,104 – 4,043,223 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -17.08 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4043104 119 - 20766785 AUAAUUGCGCGUUCAGAAAAUGAAAAUUCAAAACAAACAAAGAAAGGCAGAAAUGAAUAAGCG-AUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGUA ....((((((.((((.....)))).......((((..((..(.....((....))......).-.))..)))).............)).)))).........((((((....)))))).. ( -19.20) >DroSec_CAF1 25779 119 - 1 AUAAUUGCGCGUUCAGAAAAUGAAAAUUCAAAACAAACAGAGAAAGGCAGA-AUGAAUAAGCGGAUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGUA ....((((((.((((.....)))).......((((..((..(.....(...-..)......)...))..)))).............)).)))).........((((((....)))))).. ( -16.00) >DroSim_CAF1 27941 120 - 1 AUAAUUGCGCGUUCAGAAAAUGAAAAUUCAAAACAAACAGAGAAAGGCAGAAAUGAAUAAGCGGAUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUAUA ....((((((.((((.....)))).......((((..((..(.....((....))......)...))..)))).............)).)))).......((((((((....)))))))) ( -21.60) >DroEre_CAF1 24924 119 - 1 AUAAUUGCGCGUGCAGAAAACGAAAAUUCAAAACAAACAAAGAAAGGCGGGAAUGAAUAAGCG-AUGGGUGCUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGCA ....((((.(((.......)))......................((.((..(((.....((((-.....))))......)))..)).)))))).......((((((((....)))))))) ( -21.60) >DroYak_CAF1 28860 119 - 1 AUAAUUGCGCGUUCAGAAAAUGAAAACUCAAAACAAAGACAGAAUGGAAGGAAUGAAUAAGCG-AUGGGUGUUGUAAAAAUUAAUGGCUGAAAAGUUAAAUGUAAGUCGUAGGAUUUGCA ...(((((..(((((.....(((....))).........(.....).......)))))..)))-))..................(((((....)))))..((((((((....)))))))) ( -18.20) >consensus AUAAUUGCGCGUUCAGAAAAUGAAAAUUCAAAACAAACAAAGAAAGGCAGAAAUGAAUAAGCG_AUGGGUGUUGUAAAAAUUAAUGGCUGCAAAGUUAAAUGUAAAUCGAAGGAUUUGUA ....(((((((..((.....((...(((((.......................)))))...))..))..))).)))).......(((((....)))))..((((((((....)))))))) (-17.08 = -16.40 + -0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:23 2006