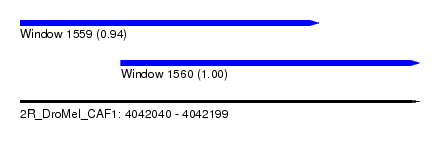
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,042,040 – 4,042,199 |
| Length | 159 |
| Max. P | 0.998714 |

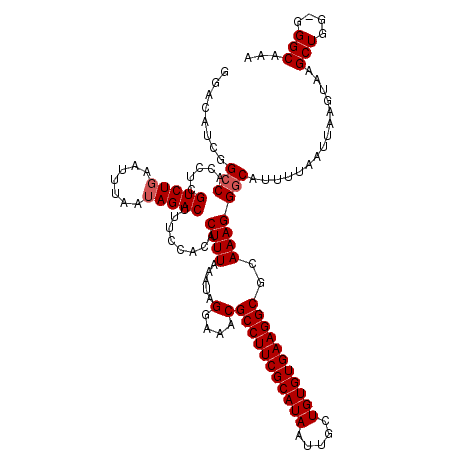
| Location | 4,042,040 – 4,042,159 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4042040 119 + 20766785 GGACAUCGGGCACCUCGUCUGAAUUUAAUAGACUUUCCACACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUUG-GGGCAAA .(...((((((.(((.(((((.......)))))(((((............)))))((((((((((((.....))))))))))))...)))..((((.....))))..)))))-)..)... ( -32.00) >DroSec_CAF1 24730 120 + 1 GGACAUCGGCCACCUCGUCUGAAUUUAAUAGACUUUCUACACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGGGGGGCAAA ........(((.(((((.((..(((((((....((((((.........)))))).((((((((((((.....))))))))))))..............))))))).)).))))))))... ( -36.70) >DroSim_CAF1 26889 120 + 1 GGACAUCGGCCACCUCGUCUGAAUUUAAUAGACUUUCCACACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGGGGGGCAAA ........(((.(((((.((..((((((((((.(.(((............(....)(((((((((((.....))))))))))).....))).).))).))))))).)).))))))))... ( -38.10) >DroEre_CAF1 23982 119 + 1 GGACUUCGGCCACCUCGUCUGGCUUUAAGAGACCUUCCGCACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGG-GGGCCAA .......((((.(((.(.((.(((((((((..((((..((..........(....)(((((((((((.....)))))))))))))..))))..)))))...)))).))).))-))))).. ( -44.60) >DroYak_CAF1 27956 115 + 1 GGACAUCGGCCACCUCGUCUGACUUUAAUAGACUUUCCACACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUA----AGCUGG-GGGCAAA ........(((.....(((((.......)))))...((.((.(((((.(((((...(((((((((((.....)))))))))))((.....)).))))).)))----)).)))-))))... ( -33.50) >consensus GGACAUCGGCCACCUCGUCUGAAUUUAAUAGACUUUCCACACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGG_GGGCAAA ........(((.....(((((.......)))))........((((.....(....)(((((((((((.....)))))))))))..)))))))...............(((....)))... (-29.30 = -29.70 + 0.40)

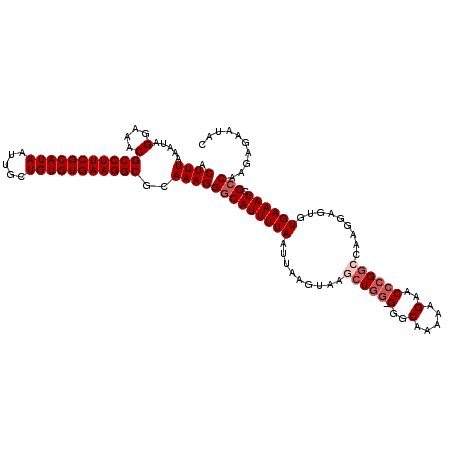
| Location | 4,042,080 – 4,042,199 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.65 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -30.41 |
| Energy contribution | -31.61 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4042080 119 + 20766785 ACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUUG-GGGCAAAAAGAACACGGCCAAGGAGUUUGAAAUCGCCAAGAGAAUAC .((((.....(....)(((((((((((.....)))))))))))..))))(((((((((((((((....))))-)(((............))).......))))))).))).......... ( -34.00) >DroSec_CAF1 24770 120 + 1 ACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGGGGGGCAAAAAGAACCCGGCCAAGGAGUGUGAAAUCGGCAAGAGAAUAC .((((.....(....)(((((((((((.....)))))))))))..))))(.(((((((.........((((((...(.....)..)))))).........)))))).).).......... ( -34.57) >DroSim_CAF1 26929 120 + 1 ACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGGGGGGCAAAAAGAACCCGGCCAAGGAGUGUGAAAUCGCCAAGAGAAUAC .((((.....(....)(((((((((((.....)))))))))))..))))(((((((((.........((((((...(.....)..)))))).........)))))).))).......... ( -39.17) >DroEre_CAF1 24022 119 + 1 ACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGG-GGGCCAACCGAACCCGCACACGGAGUGUGAAAUCGCCAAGCGUAUAC ..........(....)(((((((((((.....)))))))))))((....((((((((((((..((..((.((-(..(.....)..)))))..))..))).)))))).)))..))...... ( -37.90) >DroYak_CAF1 27996 115 + 1 ACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUA----AGCUGG-GGGCAAAAAGAGCCCGGACAAGGAGUGUGAAAUCGCCAAGAGAAUAC .((((.....(....)(((((((((((.....)))))))))))..))))(((((((((.((.----..((((-(..(.....)..))))).....))...)))))).))).......... ( -36.40) >consensus ACUUUAAAUAGGAAACGCCUUCGCAUAAUUGCUGUGUGAAGGCGCAAAGGGCAUUUUAAUUAAGUAAGCUGG_GGGCAAAAAGAACCCGGCCAAGGAGUGUGAAAUCGCCAAGAGAAUAC .((((.....(....)(((((((((((.....)))))))))))..))))(((((((((.........(((((.(..(.....)..)))))).........)))))).))).......... (-30.41 = -31.61 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:19 2006