
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,041,814 – 4,041,960 |
| Length | 146 |
| Max. P | 0.765494 |

| Location | 4,041,814 – 4,041,934 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -34.64 |
| Energy contribution | -35.08 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4041814 120 - 20766785 CAGGAGCUUUAAGGAUAUUAGGAUGGGUAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGAUGCUGGACCCUAUCAUAAAUUGAAUUCGCCCCAAAACGAA ..((.((....((((((((.((...(..((..((((((..(....)..))))))..))..).)).)))))((((.....)))).....))).(((.....)))....)).))........ ( -33.80) >DroSec_CAF1 24500 119 - 1 CGGGAGCUUUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGAUGCUGGACCCUAUCAUAAAUUGAAUUCGCCCCA-AACGAA .(((.((....((((((((.((...(((((..((((((..(....)..))))))..))))).)).)))))((((.....)))).....))).(((.....)))....))))).-...... ( -40.90) >DroSim_CAF1 26653 119 - 1 CGGGAGCUUUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGAUGCUGGACCCUAUCAUAAAUUGAAUUCGCCCCA-AACGAA .(((.((....((((((((.((...(((((..((((((..(....)..))))))..))))).)).)))))((((.....)))).....))).(((.....)))....))))).-...... ( -40.90) >DroEre_CAF1 23790 119 - 1 CGGGAGCCGUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGAUGCUGGACCCUAUCAUAAAUUGAAUUCGCCCCA-AGCGAA .(((..((....))(((((.((...(((((..((((((..(....)..))))))..))))).)).)))))((((.....))))....)))..(((.....))).(((((....-.))))) ( -42.00) >DroYak_CAF1 27719 119 - 1 CAGGAGCUUUAAGGACAUUAGAAUGGGCAGUGAGGCGUGUCCCUGAGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGAUGCUGGGCCCUAUCAUAAAUUGAAUUCGCCCCA-AACGAA ....(((.((((.(.(((.......(((((..((((((..........))))))..))))).......))))....)))).)))((((....(((.....)))....))))..-...... ( -35.64) >consensus CGGGAGCUUUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGAUGCUGGACCCUAUCAUAAAUUGAAUUCGCCCCA_AACGAA .(((.((....(((...((((....(((((..((((((..(....)..))))))..))))).))))....((((.....)))).....))).(((.....)))....)))))........ (-34.64 = -35.08 + 0.44)



| Location | 4,041,854 – 4,041,960 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -28.38 |
| Energy contribution | -29.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4041854 106 - 20766785 UGGAUGCG---GGGU------GC-----UUGGUGCGGGAGCAGGAGCUUUAAGGAUAUUAGGAUGGGUAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGA ...((((.---....------((-----((..(((....))).)))).......(((((.((...(..((..((((((..(....)..))))))..))..).)).)))))))))...... ( -34.60) >DroSec_CAF1 24539 111 - 1 CGGAUGCG---GGGU------GCGGUUGCGGGUGCGGGAGCGGGAGCUUUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGA ((.((.((---.((.------....)).)).)).))(((((....)))))..(.(((((.((...(((((..((((((..(....)..))))))..))))).)).))))).)........ ( -40.40) >DroSim_CAF1 26692 117 - 1 CGGAUGCG---GGGUGUGGGUGCGGGUGCGGGUGCGGGAGCGGGAGCUUUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGA ((.((.((---.....)).)).)).(((((.((.(((((((....))))................(((((..((((((..(....)..))))))..))))).))).)).)))))...... ( -42.20) >DroEre_CAF1 23829 87 - 1 ---------------------------------GCGGGUGCGGGAGCCGUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGA ---------------------------------(((.((.(((...((....))...........(((((..((((((..(....)..))))))..))))).))).)).)))........ ( -34.90) >DroYak_CAF1 27758 120 - 1 UGGGUGCGCAUGGGUGCGGUUGCCGGUGAGGAUGAGGGUGCAGGAGCUUUAAGGACAUUAGAAUGGGCAGUGAGGCGUGUCCCUGAGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGA ...(((((((....))).....(((((((.(..((((((.((((.((((((..(.(((....)))..)..)))))).....))))...)).))))..).)))))))....))))...... ( -39.10) >consensus CGGAUGCG___GGGU______GC_G_UGCGGGUGCGGGAGCGGGAGCUUUAAGGAUAUUAGGAUGGGCAGUGAGGCGUGGCCACGGGAACGCCUUGCUGUCACUGGAUAUGCAUGAUUGA ...............................((((((((((....))))).......((((....(((((..((((((..(....)..))))))..))))).))))...)))))...... (-28.38 = -29.50 + 1.12)

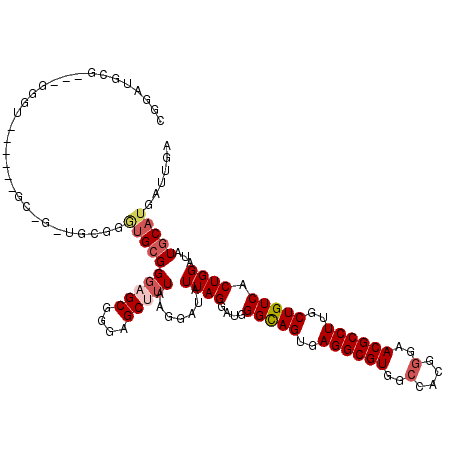
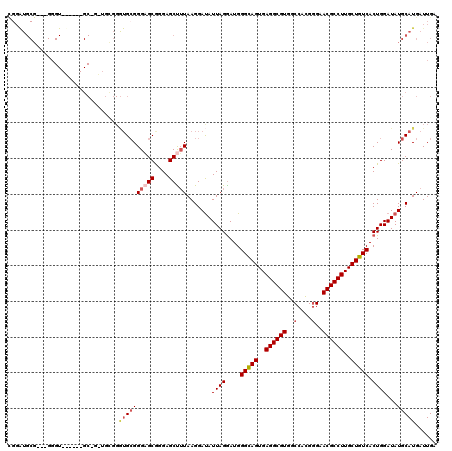
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:17 2006