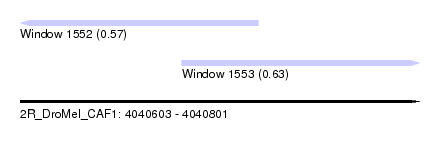
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,040,603 – 4,040,801 |
| Length | 198 |
| Max. P | 0.625254 |

| Location | 4,040,603 – 4,040,721 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -41.96 |
| Consensus MFE | -31.75 |
| Energy contribution | -31.80 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4040603 118 - 20766785 ACUCCCCCGA--UAUUGUGUGUUAGGCAAGUCAGGCCCUUGGGCCCCAUUUAAUUAGCUUGCUGGCGGUCAAGCUGGGCAUCUAUUGAAUGGCUGAUAAGCCUAGCCAACAGUGCAGCCA ........(.--((((((..(((((((..((((((((....))))(((((((((.((..(((..((......))..).)).))))))))))).))))..)))))))..)))))))..... ( -43.20) >DroSec_CAF1 23226 118 - 1 ACUCCCCCGA--UAUUGUGGGUUUGGCAAGUCGGGCCCUUGGGCCCCAUUUAAUUAGCUUGCUGGAGGUCAAGCUGGGCAUCUAUUGAAUGGUUGAUAAGCCUGGCCAACAGUGCAGCCA .....((((.--.....))))..((((((((.(((((....))))).)))).....((..((((..(((((.(((...((.((((...)))).))...))).)))))..)))))).)))) ( -43.90) >DroSim_CAF1 25377 118 - 1 ACUCCCCCGA--UACUGUGGGUUUGGCAAGUCGGGCCCUUGGGCCCCAUUUAAUUAGCUUGCUGGCGGUCAAGCUGGGCAUCUAUUGAAUGGCUGAUAAGCCUGGCCAACAGUGCAGCCA ........(.--((((((.(((..(((..((((((((....)))))((((((((.((..(((..((......))..).)).))))))))))...)))..)))..))).)))))))..... ( -45.70) >DroEre_CAF1 22671 106 - 1 ACUCCUGCAA--UACUGUGGGUCAGUCAAGUCGGACCCUUGGGUCCCCUUUAAUUAGCUAGC------------UGGGCAUCAAUUGAAUGGCCAAUAAGCCGGGCCAGUAGUGCAGCCU ....(((((.--(((((..((..(((.(((..(((((....)))))..))).)))..)).((------------(.(((....((((......))))..))).)))))))).)))))... ( -35.50) >DroYak_CAF1 26274 120 - 1 ACUCCCCCGAUGUAUUGUGGGUAAGGCAAGUCGGGCCCCAGGGCCCCGUUUAAUUAGCUAGCUGGCGGUCAGGCUGGGCAUCAAUUGAAUUGCUGAUAAGCCUAACCAGUAGUGCAGCCU ..........(((((((..(((.((((..((((((((....))))).((((((((.(((((((........)))).)))...))))))))....)))..)))).)))..))))))).... ( -41.50) >consensus ACUCCCCCGA__UAUUGUGGGUUAGGCAAGUCGGGCCCUUGGGCCCCAUUUAAUUAGCUUGCUGGCGGUCAAGCUGGGCAUCUAUUGAAUGGCUGAUAAGCCUGGCCAACAGUGCAGCCA ............((((((.((((.(((..((((((((....))))(((((((((..(((((((........))).))))....))))))))).))))..))).)))).))))))...... (-31.75 = -31.80 + 0.05)

| Location | 4,040,683 – 4,040,801 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.01 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -23.67 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4040683 118 + 20766785 AAGGGCCUGACUUGCCUAACACACAAUA--UCGGGGGAGUCGCUCAUCUGCCCAUGAAUAUAAUUAACCGCAUUAUUCAAGUUUAUCGAGUGCGCACUCCACACACGUCCUCAAUAAAUA ..((((.((((((.(((...........--..))).)))))).......)))).((((((.............)))))).((((((.(((..((...........))..))).)))))). ( -26.24) >DroSec_CAF1 23306 118 + 1 AAGGGCCCGACUUGCCAAACCCACAAUA--UCGGGGGAGUCGCUCAUCUGCCCAUGAAUAUAAUUAACCGCAUUAUUCAAGUUUAUCGAGUGCGCACUCCACACACGUCCUCAAUAAAUA ..((((.((((((.((............--...)).)))))).......)))).((((((.............)))))).((((((.(((..((...........))..))).)))))). ( -26.78) >DroSim_CAF1 25457 118 + 1 AAGGGCCCGACUUGCCAAACCCACAGUA--UCGGGGGAGUCGCUCAUCUGCCCAUGAAUAUAAUUAACCGCAUUAUUCAAGUUUAUCGAGUGCGCACUCCACACACGUCCUCAAUAAAUA ..((((.((((((.((..((.....)).--...)).)))))).......)))).((((((.............)))))).((((((.(((..((...........))..))).)))))). ( -27.32) >DroEre_CAF1 22739 117 + 1 AAGGGUCCGACUUGACUGACCCACAGUA--UUGCAGGAGUCGCUCAUCUGCCCAUGAAUAUAAUUAACCGCAUUAUUCAAGUUUAUCGAGUGCGCAC-CCACACACGUCCUCCAUAAUUA ..((((.((((((((..(((........--..((((..(.....)..))))...((((((.............)))))).)))..)))))).)).))-)).................... ( -25.72) >DroYak_CAF1 26354 120 + 1 UGGGGCCCGACUUGCCUUACCCACAAUACAUCGGGGGAGUCGCUCAUCUGCCCCUGAAUAUAAUUAACCGCAUUAUUCAAGUUUAUCGAGUGCGCGCUCCACACACGUCCUCAAUAAAUA .(((((.((((((.(((...............))).)))))).......)))))((((((.............)))))).((((((.(((..((.(.......).))..))).)))))). ( -31.08) >consensus AAGGGCCCGACUUGCCUAACCCACAAUA__UCGGGGGAGUCGCUCAUCUGCCCAUGAAUAUAAUUAACCGCAUUAUUCAAGUUUAUCGAGUGCGCACUCCACACACGUCCUCAAUAAAUA ..((((.((((((.((.................)).)))))).......)))).((((((.............)))))).((((((.(((..((...........))..))).)))))). (-23.67 = -23.95 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:12 2006