| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,338,505 – 1,338,620 |
| Length | 115 |
| Max. P | 0.898642 |

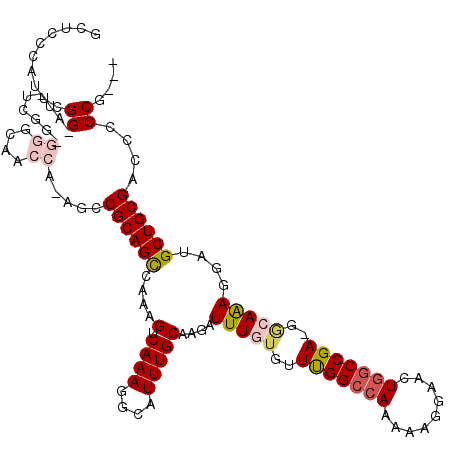
| Location | 1,338,505 – 1,338,620 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.96 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
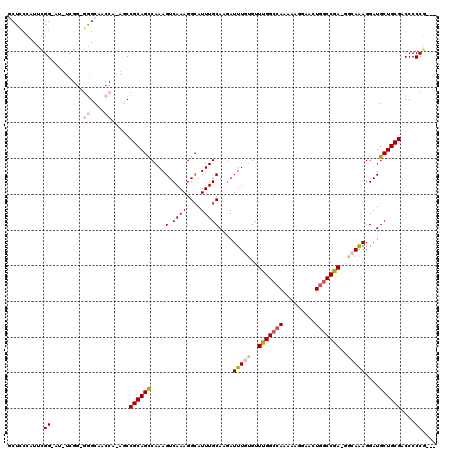
>2R_DroMel_CAF1 1338505 115 + 20766785 GCUCCCAUUCGGAAUUUCGGGGGGCAACCA-AGCCGCAGCCAAAGUCAAAGGCAUUUGCAAGAUUUGUGUUUGGCCAAAAAGGAACUGGCCGA-GGCAAAGGAUGCUGCGACCCCCG--- ..(((.....)))....((((((((.....-.))((((((....(((....((....))....(((((.((((((((.........)))))))-)))))).))))))))).))))))--- ( -40.70) >DroSec_CAF1 56941 103 + 1 GCUCCCAUUUGG---------GAGCAA-----CCCGCAGCCAAAGUCAAAGGAAUUUGCAAGAUUUGUGUUCGGCCAAAAAGGAACUGGCCGAAGGCAAAGGAUGCUGCGACCCCCG--- ((((((....))---------))))..-----..((((((((((.((....)).)))).....(((((.((((((((.........)))))))).)))))....)))))).......--- ( -43.80) >DroSim_CAF1 35157 102 + 1 GCUCCCAUUCGG---------GAGCAA-----CCCGCAGCCAAAGUCAAAGGCAUUUGCAAGAUUUGAGUUCGGCCAAAAAGGAACUGGCCGA-GCCAAAGGAUGCUGCGACCCCCG--- ((((((....))---------))))..-----..((((((....(((....((....))....((((.(((((((((.........)))))))-)))))).))))))))).......--- ( -43.10) >DroEre_CAF1 38411 119 + 1 ACUUCCCUCCGGGAUCUCGGAGGGCAACCAAAGCCGCAGCCAAAGUCAAAGGCAUUUGCAAGAUUUCUGUUUGGCCAAAAGGGAACUGGCCGA-GAAAGAGGAUGCUGCGACCCCCGGUG ....((((((((....))))))))..(((...(.((((((((((((.....)).)))).....(((((.((((((((...(....))))))))-)..)))))..)))))).)....))). ( -41.30) >DroYak_CAF1 25556 116 + 1 ACUGAUAUUCGGGAUUUCGGAGGGCAACCAAAGCCGCAGUCAAAGUCAAAGGCAUUUGCAAAAUUUAUGUUUGGUAAAAAAAGAACUGCCCGA-CGAAAAGGAUGCUGCGACCCCCA--- ..........(((.(((((..(((((......(((...............))).(((((.((((....)))).)))))........)))))..-))))).((.........))))).--- ( -28.26) >consensus GCUCCCAUUCGG_AU_UCGG_GGGCAACCA_AGCCGCAGCCAAAGUCAAAGGCAUUUGCAAGAUUUGUGUUUGGCCAAAAAGGAACUGGCCGA_GGCAAAGGAUGCUGCGACCCCCG___ ..........((.........((....)).....((((((....(.((((....)))))....(((((..(((((((.........)))))))..)))))....))))))....)).... (-21.84 = -22.96 + 1.12)

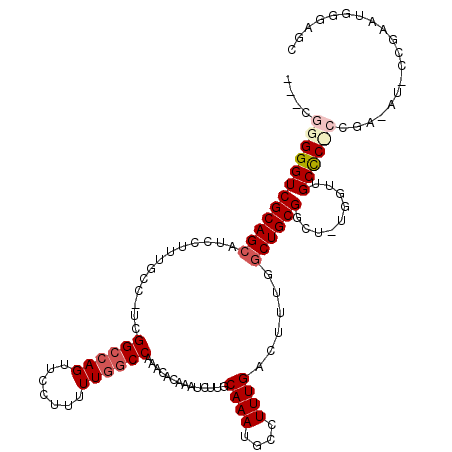
| Location | 1,338,505 – 1,338,620 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -39.25 |
| Consensus MFE | -22.10 |
| Energy contribution | -23.18 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1338505 115 - 20766785 ---CGGGGGUCGCAGCAUCCUUUGCC-UCGGCCAGUUCCUUUUUGGCCAAACACAAAUCUUGCAAAUGCCUUUGACUUUGGCUGCGGCU-UGGUUGCCCCCCGAAAUUCCGAAUGGGAGC ---((((((..(((((.......(((-..((((((.......)))))).............(((...(((.........))))))))).-..)))))))))))...((((.....)))). ( -41.70) >DroSec_CAF1 56941 103 - 1 ---CGGGGGUCGCAGCAUCCUUUGCCUUCGGCCAGUUCCUUUUUGGCCGAACACAAAUCUUGCAAAUUCCUUUGACUUUGGCUGCGGG-----UUGCUC---------CCAAAUGGGAGC ---...((.(((((((....((((..(((((((((.......)))))))))..)))).....((((.((....)).))))))))))).-----))((((---------((....)))))) ( -41.60) >DroSim_CAF1 35157 102 - 1 ---CGGGGGUCGCAGCAUCCUUUGGC-UCGGCCAGUUCCUUUUUGGCCGAACUCAAAUCUUGCAAAUGCCUUUGACUUUGGCUGCGGG-----UUGCUC---------CCGAAUGGGAGC ---...((.((((((((...(((((.-((((((((.......)))))))).).))))...)))....(((.........)))))))).-----))((((---------((....)))))) ( -39.90) >DroEre_CAF1 38411 119 - 1 CACCGGGGGUCGCAGCAUCCUCUUUC-UCGGCCAGUUCCCUUUUGGCCAAACAGAAAUCUUGCAAAUGCCUUUGACUUUGGCUGCGGCUUUGGUUGCCCUCCGAGAUCCCGGAGGGAAGU .(((((((.(((((((..........-..((((((.......))))))...((((..((..((....))....)).))))))))))))))))))..(((((((......))))))).... ( -46.40) >DroYak_CAF1 25556 116 - 1 ---UGGGGGUCGCAGCAUCCUUUUCG-UCGGGCAGUUCUUUUUUUACCAAACAUAAAUUUUGCAAAUGCCUUUGACUUUGACUGCGGCUUUGGUUGCCCUCCGAAAUCCCGAAUAUCAGU ---.(((((((((((((......(((-..(((((........((((.......)))).........))))).)))...)).)))))))(((((.......))))).)))).......... ( -26.63) >consensus ___CGGGGGUCGCAGCAUCCUUUGCC_UCGGCCAGUUCCUUUUUGGCCAAACACAAAUCUUGCAAAUGCCUUUGACUUUGGCUGCGGCU_UGGUUGCCC_CCGA_AU_CCGAAUGGGAGC ....((((((((((((.............((((((.......))))))..............((((....))))......)))))).........))))))................... (-22.10 = -23.18 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:28 2006