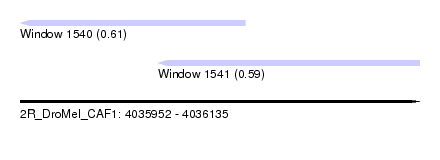
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,035,952 – 4,036,135 |
| Length | 183 |
| Max. P | 0.612235 |

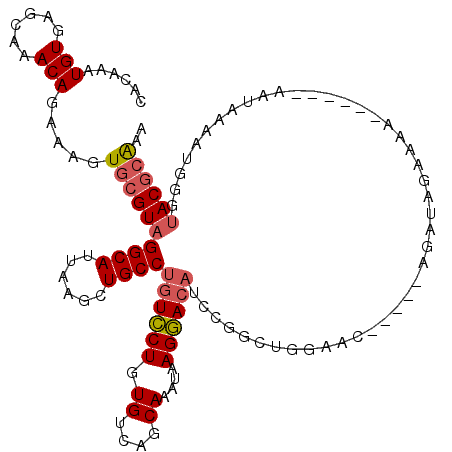
| Location | 4,035,952 – 4,036,055 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4035952 103 - 20766785 CACAAAUGUGGGCAAACAGAAAGUGCGUAGGCAUUAAGCUGCCUGUCCUGUGUCAGCAAAUAAGGACAUCCGGCUGGAAA-----AGAUAGAAA------------AAUGGAUACGCAAA (((....))).............((((((..((((.(((((..((((((.((....))....))))))..))))).....-----.........------------))))..)))))).. ( -26.76) >DroSec_CAF1 18609 115 - 1 CACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAAGCUGCCUGUCCUGUGUCAGCAAAUAAGGACAUCCGGCUGGAAC-----AGAUAGAAAAAAAUGAAAUAAAAUGGUUACGCAAA (((....))).............(((((((.((((.(((((..((((((.((....))....))))))..)))))....(-----(............))......)))).))))))).. ( -29.40) >DroSim_CAF1 20740 115 - 1 CACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAAGCUGCCUGUCCUGUGUCACCAAAUAAGGACAUCCGGCUGGAAC-----AGAUAGAAAAAAAUGAAAUAAAAUGGUUACGCAAA (((....))).............(((((((.((((.(((((..((((((.((....))....))))))..)))))....(-----(............))......)))).))))))).. ( -28.20) >DroEre_CAF1 17790 103 - 1 ------UGUGAGCAAACAGAAAGUGCGUAGGCAUUAAUCUGCCUGUCCUGUGUCAGCAAAUAAGGACAUCCGGCUCGAAC-----AGAAAGAAAA------AAUAAAAUGGGCACCAGGA ------..(((((...((((.(((((....)))))..))))..((((((.((....))....))))))....)))))...-----..........------.......(((...)))... ( -22.80) >DroYak_CAF1 21856 114 - 1 CACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAGGCUGCCUCUUCUGUGUCAGCAAAUAAGAACCUCCGGAUUGUAUCUGUUAUUUGAAAAU------AAUGAAAUGGGUACGCAGA (((....))).((........(((.((.(((..(((.((((.(.(....).).))))...)))...))).)).)))((((((.(((((.......------)))))...))))))))... ( -23.90) >consensus CACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAAGCUGCCUGUCCUGUGUCAGCAAAUAAGGACAUCCGGCUGGAAC_____AGAUAGAAAA______AAUAAAAUGGGUACGCAAA ......(((......))).....((((((((((......))))((((((.((....))....))))))............................................)))))).. (-17.98 = -18.50 + 0.52)

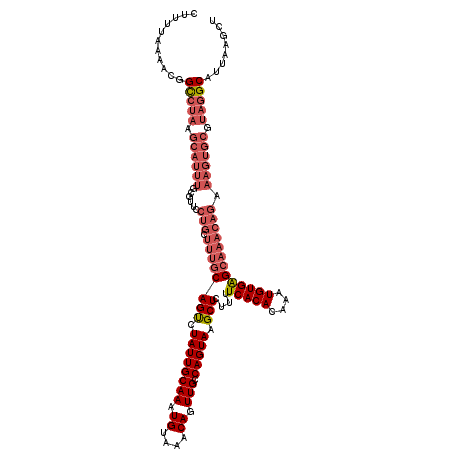
| Location | 4,036,015 – 4,036,135 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -20.70 |
| Energy contribution | -24.07 |
| Covariance contribution | 3.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4036015 120 - 20766785 CUUUUAAAACGGUCUAAGCAUUUGCGUUCCUGCUUUGCAGUCUAUUGCAAAUGUAAACAGUUGCCAGUAAGCUCUUUUCACACAAAUGUGGGCAAACAGAAAGUGCGUAGGCAUUAAGCU ...........(((((.((((((......(((.(((((((..((((((((.((....)).))).)))))..))....(((((....))))))))))))).)))))).)))))........ ( -29.90) >DroSec_CAF1 18684 120 - 1 CUUUUAAAACGGCCUAAGCAUUUGCGUUCCUGCUUUGCAGCCUAUUGCAAAUGUAAACAGUUGCCAGUAAGCUCUUUUCACACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAAGCU ...........(((((.((((((......(((.((((((((.((((((((.((....)).))).))))).)))....(((((....))))))))))))).)))))).)))))........ ( -34.80) >DroSim_CAF1 20815 120 - 1 CUUUUAAAACGGCCUAAGCAUUUGCGUUCCUGCUUUCCAGUCUAUUGCAAAUGUAAACAGUUGCCAGUAAGCUCUUUUCACACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAAGCU ...........(((((.((((((..(((..(((.....((..((((((((.((....)).))).)))))..))....(((((....)))))))))))...)))))).)))))........ ( -30.50) >DroYak_CAF1 21930 98 - 1 CUUUUAAAACGGU----------------------CGCAGUCUAUUGCAAAUGUAAACAGUUGCCAGUAAGCUCUUUUCACACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAGGCU ..........(((----------------------(((((..((((((((.((....)).))).)))))..))....(((((....)))))))........(((((....))))).)))) ( -21.60) >consensus CUUUUAAAACGGCCUAAGCAUUUGCGUUCCUGCUUUGCAGUCUAUUGCAAAUGUAAACAGUUGCCAGUAAGCUCUUUUCACACAAAUGUGAGCAAACAGAAAGUGCGUAGGCAUUAAGCU ...........(((((.((((((......(((.((((((((.((((((((.((....)).))).))))).)))....(((((....))))))))))))).)))))).)))))........ (-20.70 = -24.07 + 3.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:00 2006