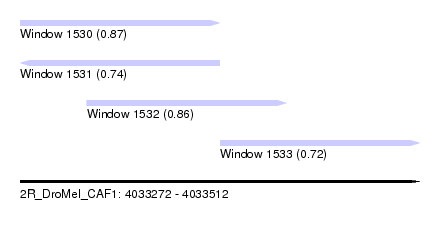
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,033,272 – 4,033,512 |
| Length | 240 |
| Max. P | 0.872800 |

| Location | 4,033,272 – 4,033,392 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -50.38 |
| Consensus MFE | -39.40 |
| Energy contribution | -40.16 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4033272 120 + 20766785 CGUGGGCACCGCCUUGCCACCAAUCCUGGUCGCCGUGCUCGUUAUUGCCGGCGAUAUGGUCGGUGCUGCUGGCGCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGC ....((((.((((..((((((.(((...(((((((.((........)))))))))..))).))))..)).)))).)))).(((((.((((....)))).)))))((((((....)))))) ( -55.30) >DroSec_CAF1 15923 120 + 1 CGUGGGCACCGCCUUGCCACCAAUCCUGGUCGCCGUGCUCGUUAGUGCCGGCGAUAUGGUCGGUGCGGAUGGACCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGC .((((.((((((((.(((((((....)))((((((.((.(....).))))))))..)))).)).))))...(((((((((......))))))..))).....)).))))(((.....))) ( -53.70) >DroSim_CAF1 18067 120 + 1 CGUGGGCACCGCCUUGCCACCAAUCCUGGUGGCCGUGCUCGCUAGUGCCGGCGAUAUGGUCGGUGCGGCUGGCGCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGC .((((((((......(((((((....))))))).))))))))..(..(((((......)))))..)(((..(((((((((......))))))).......))..)))..(((.....))) ( -59.31) >DroEre_CAF1 15274 120 + 1 CGUGGGCACCGACUUGCCACCAAUCCUGGUCUCCGUGCUCGUUAGUGCCGGCGAUAUGGUCGGUGCGGCUGGCACUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGC .(((((((((((((((((((((....))))....(..((....))..).))))....)))))))))...(((((((((((......)))))).))))).......))))(((.....))) ( -52.10) >DroYak_CAF1 19022 105 + 1 CGUGGGCACCGCCUUACCACCGAUCCUGCUCUCCGUGCUCGUUAGUGUUGGCAAUAUGGUCGGU---------------ACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGC .((((((...))).))).(((((((.((((....(..((....))..).))))....)))))))---------------.(((((.((((....)))).)))))((((((....)))))) ( -31.50) >consensus CGUGGGCACCGCCUUGCCACCAAUCCUGGUCGCCGUGCUCGUUAGUGCCGGCGAUAUGGUCGGUGCGGCUGGCGCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGC .((((((((((....(((((((....)))((((((.((........))))))))..))))))))))..............(((((.((((....)))).))))).))))(((.....))) (-39.40 = -40.16 + 0.76)

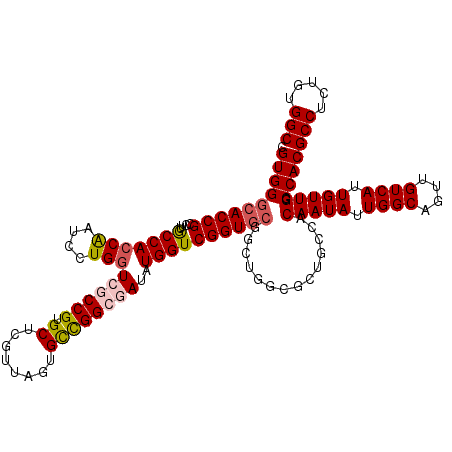
| Location | 4,033,272 – 4,033,392 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -34.24 |
| Energy contribution | -35.86 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4033272 120 - 20766785 GCCACAGAGGCGUGGCCAACAAUGACAACUGCCAAUAUUGUGGCAGCGCCAGCAGCACCGACCAUAUCGCCGGCAAUAACGAGCACGGCGACCAGGAUUGGUGGCAAGGCGGUGCCCACG (((.....)))((((.((..........((((((......))))))((((.((..((((((((...((((((((........)).))))))...)).))))))))..)))).)).)))). ( -51.10) >DroSec_CAF1 15923 120 - 1 GCCACAGAGGCGUGGCCAACAAUGACAACUGCCAAUAUUGUGGCAGGUCCAUCCGCACCGACCAUAUCGCCGGCACUAACGAGCACGGCGACCAGGAUUGGUGGCAAGGCGGUGCCCACG (((.....)))((((........(((..((((((......))))))))).....((((((.((....(((((((.(....).)).)))))(((......))).....)))))))))))). ( -50.70) >DroSim_CAF1 18067 120 - 1 GCCACAGAGGCGUGGCCAACAAUGACAACUGCCAAUAUUGUGGCAGCGCCAGCCGCACCGACCAUAUCGCCGGCACUAGCGAGCACGGCCACCAGGAUUGGUGGCAAGGCGGUGCCCACG ........(((.((((((....))....((((((......)))))).)))))))((((((.((...((((........)))).....((((((......))))))..))))))))..... ( -50.50) >DroEre_CAF1 15274 120 - 1 GCCACAGAGGCGUGGCCAACAAUGACAACUGCCAAUAUUGUGGCAGUGCCAGCCGCACCGACCAUAUCGCCGGCACUAACGAGCACGGAGACCAGGAUUGGUGGCAAGUCGGUGCCCACG (((.....)))((((.((....((((.(((((((......)))))))....(((..(((((((...((.(((((.(....).)).))).))...)).))))))))..)))).)).)))). ( -48.00) >DroYak_CAF1 19022 105 - 1 GCCACAGAGGCGUGGCCAACAAUGACAACUGCCAAUAUUGU---------------ACCGACCAUAUUGCCAACACUAACGAGCACGGAGAGCAGGAUCGGUGGUAAGGCGGUGCCCACG (((.....)))((((.......((........)).....((---------------((((.((.(((..((....(....).((.......))......))..))).)))))))))))). ( -30.20) >consensus GCCACAGAGGCGUGGCCAACAAUGACAACUGCCAAUAUUGUGGCAGCGCCAGCCGCACCGACCAUAUCGCCGGCACUAACGAGCACGGCGACCAGGAUUGGUGGCAAGGCGGUGCCCACG (((((......)))))......((...((((((.......(((.....)))....((((((((...((((((((.(....).)).))))))...)).))))))....))))))...)).. (-34.24 = -35.86 + 1.62)

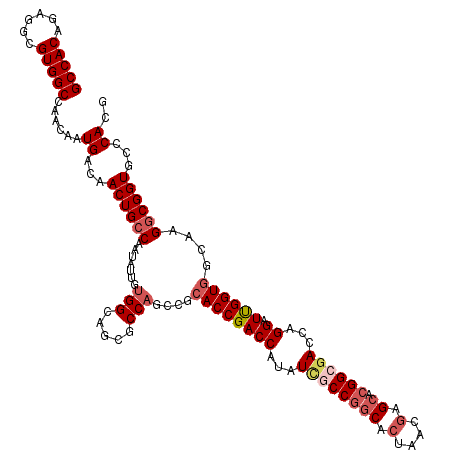
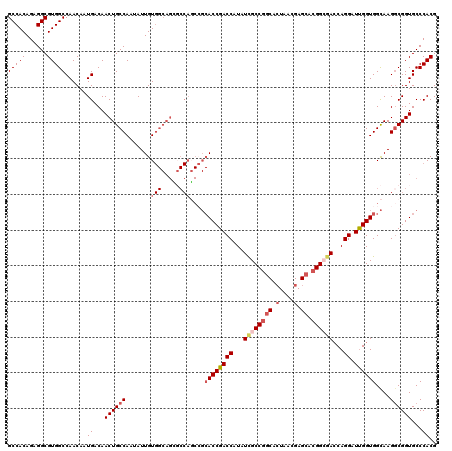
| Location | 4,033,312 – 4,033,432 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -44.34 |
| Consensus MFE | -35.62 |
| Energy contribution | -36.80 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4033312 120 + 20766785 GUUAUUGCCGGCGAUAUGGUCGGUGCUGCUGGCGCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGCAUUUGCAUAGCCAUGGGUUAGUUCGGCAUCCGGAUUGAAU .........((((...(((.((((((.....)))))))))(((((.((((....)))).)))))....))))((((((((.........))))))))(((((((((...))))))))).. ( -44.60) >DroSec_CAF1 15963 120 + 1 GUUAGUGCCGGCGAUAUGGUCGGUGCGGAUGGACCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGCAUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUGAAU ....(..(((((......)))))..)(((((..(((((((((.....(((.((.((((....)))).)))))..)))))))...((((((((...)))).)))))))))))......... ( -47.20) >DroSim_CAF1 18107 120 + 1 GCUAGUGCCGGCGAUAUGGUCGGUGCGGCUGGCGCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGCAUUUGCAUAGCCAUGGGUUAGUGCGGCGUGCUCAUUGAAU (((.(..(((((......)))))..))))(((((((((((......)))))).)))))....((..((((((.(((((((.........)))))))((....))))))))..))...... ( -50.40) >DroEre_CAF1 15314 120 + 1 GUUAGUGCCGGCGAUAUGGUCGGUGCGGCUGGCACUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGCAUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUCAAU ....(..(((((......)))))..)((((((((.(((((((.....(((.((.((((....)))).)))))..)))))))..))).))))).((((((....(((...))))))))).. ( -47.40) >DroYak_CAF1 19062 105 + 1 GUUAGUGUUGGCAAUAUGGUCGGU---------------ACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGCAUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUGAAU (((.(..(((((..((((((..((---------------((((((.((((....)))).)))))((((((....))))))...)))...)))))).)))))..))))............. ( -32.10) >consensus GUUAGUGCCGGCGAUAUGGUCGGUGCGGCUGGCGCUGCCACAAUAUUGGCAGUUGUCAUUGUUGGCCACGCCUCUGUGGCAUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUGAAU ....((((((((((..(((.((((((.....)))))))))(((((.((((....)))).)))))((((((....))))))..)))).(((((...)))))...))))))........... (-35.62 = -36.80 + 1.18)

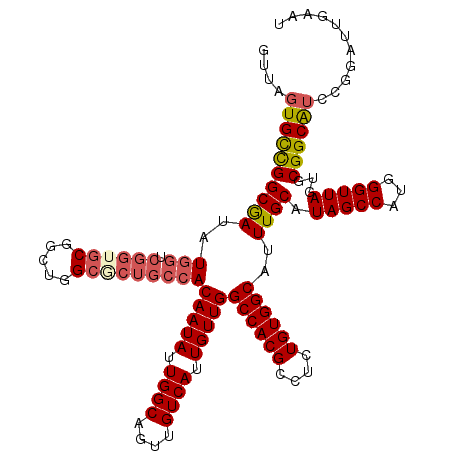
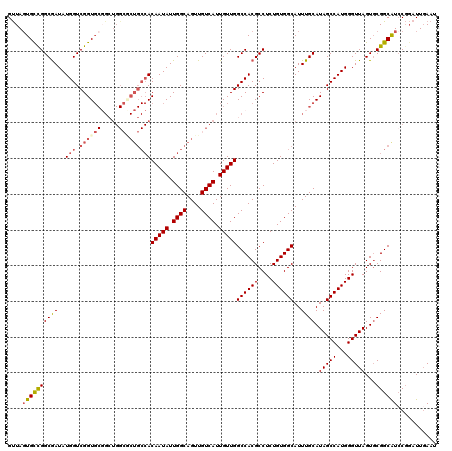
| Location | 4,033,392 – 4,033,512 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -27.83 |
| Energy contribution | -27.91 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4033392 120 + 20766785 AUUUGCAUAGCCAUGGGUUAGUUCGGCAUCCGGAUUGAAUUUUAUAUCGUCCCUCUGAACACUCUGGCCAAUCUUAUUGGUCUUGUCCGACUCGCUGCGCUUUGUUUUUGCUAGUAUUAU ....(((.(((((((((((.((((((.....((((.((........))))))..)))))).....(((((((...)))))))......)))))).)).))).)))............... ( -27.40) >DroSec_CAF1 16043 120 + 1 AUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUGAAUUUUAUAUCGUCCCUCUGAACACUCUGGCCAAUCUUAUUGGUCUUGUCCGACUCGCUGCGCUUUGUUUUUGCUGGCAAUAU ..((((.((((........((((((((...(((((...........(((......))).......(((((((...)))))))..)))))....))))))))........))))))))... ( -32.49) >DroSim_CAF1 18187 120 + 1 AUUUGCAUAGCCAUGGGUUAGUGCGGCGUGCUCAUUGAAUUUUAUAUCGUACCUCUGAACACUCUGGCCAAUCUUAUUGGUCUUGUCCGACUCGCCGCGCUUUGUUUUUGCUGGCAACAU ..((((.((((........(((((((((((.(((..(...............)..))).)))...(((((((...)))))))...........))))))))........))))))))... ( -33.55) >DroEre_CAF1 15394 120 + 1 AUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUCAAUUUCAUUUCGUCCCUCUGAACACUCAGGCCAAUCUUAUUGGUCUUUUCCGACCCGCUGCGCUUUGUUUUUGCUGGCUUUAU ....((.((((........((((((((...((((...........((((......)))).....((((((((...))))))))..))))....))))))))........))))))..... ( -30.79) >DroYak_CAF1 19127 120 + 1 AUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUGAAUUUUAUUUCGUCCCUCCGAACACUCAGGCCAAUCUUAUUGGUCUUUUCCGACCCGCUGCGCUUUGUUUUUGCUGGUAUUAU ...(((.((((........((((((((...((((.((((......))))....)))).......((((((((...))))))))..........))))))))........))))))).... ( -32.39) >consensus AUUUGCAUAGCCAUGGGUUAGUGCGGCAUCCGGAUUGAAUUUUAUAUCGUCCCUCUGAACACUCUGGCCAAUCUUAUUGGUCUUGUCCGACUCGCUGCGCUUUGUUUUUGCUGGCAUUAU ...(((.((((........((((((((...((((............(((......))).......(((((((...)))))))...))))....))))))))........))))))).... (-27.83 = -27.91 + 0.08)

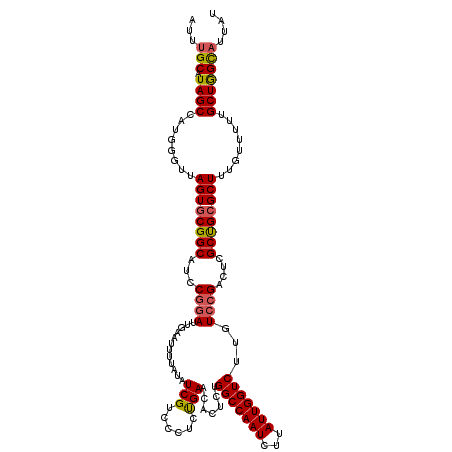
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:52 2006