| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,030,055 – 4,030,215 |
| Length | 160 |
| Max. P | 0.894482 |

| Location | 4,030,055 – 4,030,175 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.76 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4030055 120 + 20766785 AAACUCAAGUGAAAGCCCAACCGAAAUGGGAAUGAAUAUAAUUCCCUGCUAAGCCAGCUUAAAGGAACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCAAGCCAGAAAUCCUGGUU ........((....))...........((((((.......))))))((((..(((.(.((((...........)))).).)))..((....))......))))((((((.....)))))) ( -24.20) >DroSec_CAF1 12778 120 + 1 AAACUCGAGUGAAAGCCCAGCCGAAAUGGGAAUGAACAUAAUUCCCCGCUUAGCCAGCUUAGAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGAAAGCCAGAAAUCCUGGUU ......(.((....)).).(((.....((((((.......))))))......(((........)))..............)))..((....))..........((((((.....)))))) ( -27.80) >DroSim_CAF1 14862 120 + 1 AAACUCGAGUGAAAGCCCAGCCGAAAUGGGAAUGAACAUAAUUCCCCGCUUAGCCAGCUUAGAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCAAGCCAGAAAUCCUGGUU ..............((...(((.....((((((.......))))))......(((........)))..............)))..((....)).......)).((((((.....)))))) ( -28.10) >DroEre_CAF1 11984 120 + 1 AAACUCGAGUGAAAACCCAGCCGAAAUGGGAAUGAAUAUAAUUCCCUGCUUAGCCAGCUUAAAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCUCGCCAGAAAUCCUGGUU ......((((.........(((.....((((((.......))))))......(((........)))..............)))..((....)).......))))(((((.....))))). ( -31.20) >DroYak_CAF1 15067 120 + 1 AAACUGGAGUGAAGACCUAGCCGAAAUGGGAAUGAAUAUAAUUCCCUGCUUAGCCAGCUUAAAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCACGGCAGAAAUCCUGGUU .(((((((((....))...((((....((((((.......))))))(((((.(((........)))...................((....)).....))))))))).....))).)))) ( -29.70) >consensus AAACUCGAGUGAAAGCCCAGCCGAAAUGGGAAUGAAUAUAAUUCCCUGCUUAGCCAGCUUAAAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCAAGCCAGAAAUCCUGGUU ........((.........(((.....((((((.......))))))......(((........)))..............)))..((....))..)).......(((((.....))))). (-22.16 = -22.76 + 0.60)

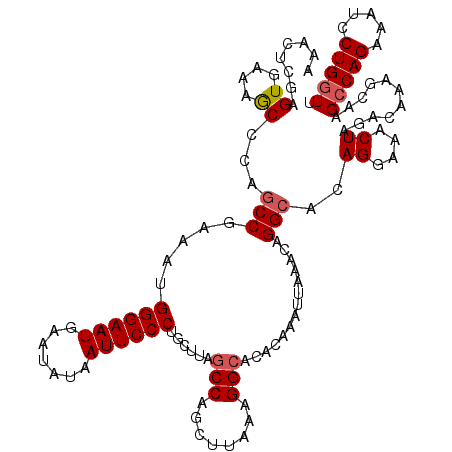
| Location | 4,030,055 – 4,030,175 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -27.62 |
| Energy contribution | -27.54 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4030055 120 - 20766785 AACCAGGAUUUCUGGCUUGCUUUGUCUAGUUUCCUGUGCCUGUUUAAUUUGUGUUCCUUUAAGCUGGCUUAGCAGGGAAUUAUAUUCAUUCCCAUUUCGGUUGGGCUUUCACUUGAGUUU ..((((.....))))...((((.((.((((((((((((((.((((((...........)))))).)))...)))))))))))........((((.......)))).....))..)))).. ( -30.10) >DroSec_CAF1 12778 120 - 1 AACCAGGAUUUCUGGCUUUCUUUGUCUAGUUUCCUGUGCCUGUUUAAUUUGUGUGCCUCUAAGCUGGCUAAGCGGGGAAUUAUGUUCAUUCCCAUUUCGGCUGGGCUUUCACUCGAGUUU ...(((((...(((((.......).))))..))))).........((((((.(((.....((((..(((.....((((((.......)))))).....)))..).))).))).)))))). ( -31.50) >DroSim_CAF1 14862 120 - 1 AACCAGGAUUUCUGGCUUGCUUUGUCUAGUUUCCUGUGCCUGUUUAAUUUGUGUGCCUCUAAGCUGGCUAAGCGGGGAAUUAUGUUCAUUCCCAUUUCGGCUGGGCUUUCACUCGAGUUU ...(((((...(((((.......).))))..))))).........((((((.(((.....((((..(((.....((((((.......)))))).....)))..).))).))).)))))). ( -31.50) >DroEre_CAF1 11984 120 - 1 AACCAGGAUUUCUGGCGAGCUUUGUCUAGUUUCCUGUGCCUGUUUAAUUUGUGUGCCUUUAAGCUGGCUAAGCAGGGAAUUAUAUUCAUUCCCAUUUCGGCUGGGUUUUCACUCGAGUUU ..((((.....))))((((.......((((((((((((((.((((((...........)))))).)))...)))))))))))........((((.......))))......))))..... ( -32.80) >DroYak_CAF1 15067 120 - 1 AACCAGGAUUUCUGCCGUGCUUUGUCUAGUUUCCUGUGCCUGUUUAAUUUGUGUGCCUUUAAGCUGGCUAAGCAGGGAAUUAUAUUCAUUCCCAUUUCGGCUAGGUCUUCACUCCAGUUU ....(((((((..(((((((((.(.(((((((...(..((..........).)..)....)))))))).)))))((((((.......))))))....)))).)))))))........... ( -30.90) >consensus AACCAGGAUUUCUGGCUUGCUUUGUCUAGUUUCCUGUGCCUGUUUAAUUUGUGUGCCUUUAAGCUGGCUAAGCAGGGAAUUAUAUUCAUUCCCAUUUCGGCUGGGCUUUCACUCGAGUUU ...(((((...(((((.......).))))..)))))..............(((.((((...(((((((...)).((((((.......))))))....)))))))))...)))........ (-27.62 = -27.54 + -0.08)

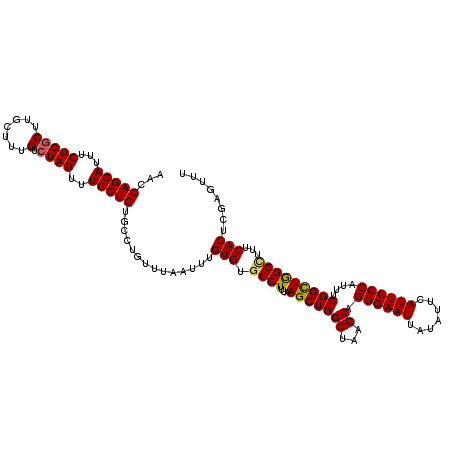
| Location | 4,030,095 – 4,030,215 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -20.50 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4030095 120 + 20766785 AUUCCCUGCUAAGCCAGCUUAAAGGAACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCAAGCCAGAAAUCCUGGUUAAAAGUCAUAACACACAGGCACGCACAAAGCCGCAUUUGU ....(((..((((....)))).)))...((((((......(((..((....)).......((.((((((.....))))))....(((..........)))..)).....)))..)))))) ( -25.90) >DroSec_CAF1 12818 120 + 1 AUUCCCCGCUUAGCCAGCUUAGAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGAAAGCCAGAAAUCCUGGUUAAAAGUCAAAACACACAGGCACGCACAAAGCCGCAUAUGU .......((...(((........)))..............(((..((....)).(((......((((((.....))))))....)))......................)))))...... ( -23.90) >DroSim_CAF1 14902 120 + 1 AUUCCCCGCUUAGCCAGCUUAGAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCAAGCCAGAAAUCCUGGUUAAAAGUCAAAACACACAGGCACGCACAAAGCCGCAUAUGU .......((...(((........)))..............(((..((....)).......((.((((((.....))))))....(((..........)))..)).....)))))...... ( -25.70) >DroEre_CAF1 12024 120 + 1 AUUCCCUGCUUAGCCAGCUUAAAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCUCGCCAGAAAUCCUGGUUAAAAGUCAAAACACACAGGCACGCACAAAGCCGCAUAUGU ......(((...(((........)))..............(((..((....)).......((..(((((.....))))).....(((..........)))..)).....))))))..... ( -24.30) >DroYak_CAF1 15107 120 + 1 AUUCCCUGCUUAGCCAGCUUAAAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCACGGCAGAAAUCCUGGUUAAAAGUCAAAACACACAGGCACGCACAAAGCCGCAUAUGU ......(((...(((........)))..............(((..((....)).(((..(((.(((.(....).))))))....)))......................))))))..... ( -20.70) >consensus AUUCCCUGCUUAGCCAGCUUAAAGGCACACAAAUUAAACAGGCACAGGAAACUAGACAAAGCAAGCCAGAAAUCCUGGUUAAAAGUCAAAACACACAGGCACGCACAAAGCCGCAUAUGU .......((...(((........)))..............(((..((....)).(((......((((((.....))))))....)))......................)))))...... (-20.50 = -21.30 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:38 2006