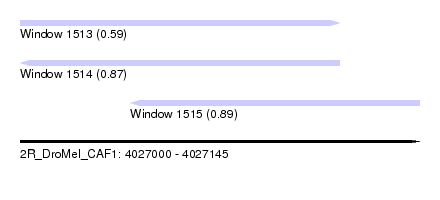
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,027,000 – 4,027,145 |
| Length | 145 |
| Max. P | 0.892378 |

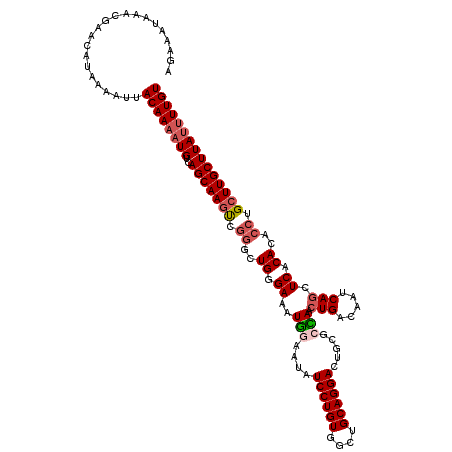
| Location | 4,027,000 – 4,027,116 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -23.89 |
| Energy contribution | -25.20 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4027000 116 + 20766785 AGGAAUAAACGAACAUAAAAUUACAAAAUGUCAGCAAGUC----UGUGAAAUGAAAUAUCCUGUGGCUGCAGGACUGCGGUAACUGACAAUCAGCUCACACACCUGCUUGCUUAUUUUGU ......................((((((((..(((((((.----(((((..(((....((..((.((((((....)))))).)).))...)))..))))).....))))))))))))))) ( -29.20) >DroSec_CAF1 9794 120 + 1 AGAAAUAAACGAACAUAAAAUUACAAAAUGUCAGCAAGUCGGGCUGGGAAAUGGAAUAUCCUGUGGCUGCAGGACUGCGCCAACUGACAAUCAGCUCACACACCUGCUUGCUUAUUUUGU ......................((((((((..((((((.((((.((((...(((..((((((((....)))))).))..))).(((.....)))))))....)))))))))))))))))) ( -33.30) >DroSim_CAF1 11858 120 + 1 AGAAAUAAACGAACAUAAAAUUACAAAAUGUCAGCAAGUCGGGCUGGGAAAUGGAAUAUCCUGUGGCUGCAGGACUGCGCCAACUGACAAUCAGCUCACACACCUGCUUGCUUAUUUUGU ......................((((((((..((((((.((((.((((...(((..((((((((....)))))).))..))).(((.....)))))))....)))))))))))))))))) ( -33.30) >DroEre_CAF1 8914 120 + 1 AGGAAUAAACGAGCAUAAAAUUACAAAAUGUCAGCAAGCCGGGCUGGGAAAUUGAAUAUCCUGUGGCUGCAGGACUGCGGGAAAUGACAAUCAGCUCGCACACCUGAUUGCUUACAUUGU ..........................(((((.(((((..((((.((............((((((....)))))).((((((...((.....)).)))))))))))).))))).))))).. ( -33.50) >consensus AGAAAUAAACGAACAUAAAAUUACAAAAUGUCAGCAAGUCGGGCUGGGAAAUGGAAUAUCCUGUGGCUGCAGGACUGCGCCAACUGACAAUCAGCUCACACACCUGCUUGCUUAUUUUGU ......................((((((((..(((((((.((..((.((..(((....((((((....)))))).....))).(((.....))).)).))..)).))))))))))))))) (-23.89 = -25.20 + 1.31)

| Location | 4,027,000 – 4,027,116 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -27.71 |
| Energy contribution | -29.53 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4027000 116 - 20766785 ACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUACCGCAGUCCUGCAGCCACAGGAUAUUUCAUUUCACA----GACUUGCUGACAUUUUGUAAUUUUAUGUUCGUUUAUUCCU ....(((((((.(((((((.(((((((((.....)))).......((((((......))))))........)))))----.)))))))(((((...........))))).)))))))... ( -34.00) >DroSec_CAF1 9794 120 - 1 ACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUGGCGCAGUCCUGCAGCCACAGGAUAUUCCAUUUCCCAGCCCGACUUGCUGACAUUUUGUAAUUUUAUGUUCGUUUAUUUCU (((((((.(((((((.(((((((.(((((.....))))).)))).((((((......)))))).............))).).))))))...)))))))...................... ( -36.60) >DroSim_CAF1 11858 120 - 1 ACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUGGCGCAGUCCUGCAGCCACAGGAUAUUCCAUUUCCCAGCCCGACUUGCUGACAUUUUGUAAUUUUAUGUUCGUUUAUUUCU (((((((.(((((((.(((((((.(((((.....))))).)))).((((((......)))))).............))).).))))))...)))))))...................... ( -36.60) >DroEre_CAF1 8914 120 - 1 ACAAUGUAAGCAAUCAGGUGUGCGAGCUGAUUGUCAUUUCCCGCAGUCCUGCAGCCACAGGAUAUUCAAUUUCCCAGCCCGGCUUGCUGACAUUUUGUAAUUUUAUGCUCGUUUAUUCCU ..(((((.(((((((.((((((.(((.((.....)).))).))).((((((......)))))).............))).)).))))).))))).......................... ( -25.90) >consensus ACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUGCCGCAGUCCUGCAGCCACAGGAUAUUCCAUUUCCCAGCCCGACUUGCUGACAUUUUGUAAUUUUAUGUUCGUUUAUUCCU (((((((.(((((((.(((((((.(((((.....))))).)))).((((((......)))))).............))).).))))))...)))))))...................... (-27.71 = -29.53 + 1.81)

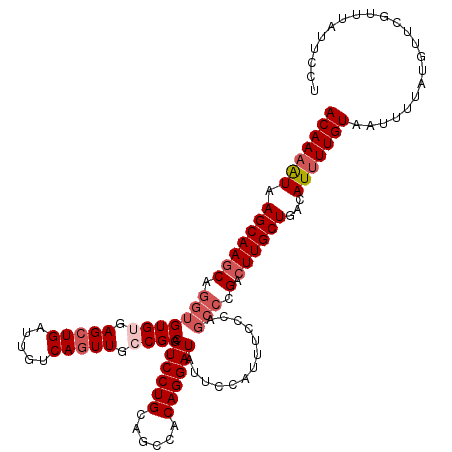
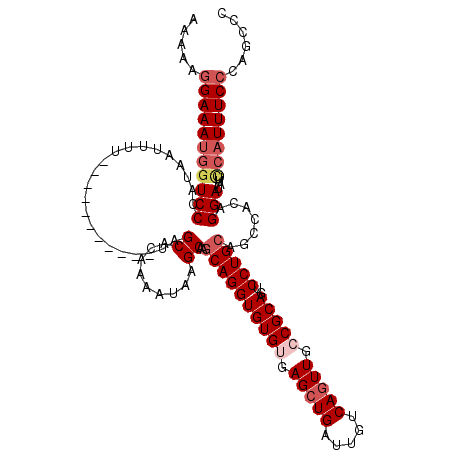
| Location | 4,027,040 – 4,027,145 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -25.30 |
| Energy contribution | -27.17 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4027040 105 - 20766785 AAAAAGGAAAUGGUCCCAUAAUUUU-----------AAGCACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUACCGCAGUCCUGCAGCCACAGGAUAUUUCAUUUCACA---- ......((((((((((.........-----------..((.........))..(((((((((..(((((.....)))))..))))..))))).......))))....))))))...---- ( -27.30) >DroSec_CAF1 9834 109 - 1 AAAAAGGAAAUGGUCCCAUAAUUUU-----------UAGCACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUGGCGCAGUCCUGCAGCCACAGGAUAUUCCAUUUCCCAGCCC .....(((((((((((.........-----------..((.........))..((((((((((.(((((.....))))).)))))..))))).......)))....))))))))...... ( -35.60) >DroSim_CAF1 11898 109 - 1 AAAAAGGAAAUGGUCCCAUAAUUUU-----------UAGCACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUGGCGCAGUCCUGCAGCCACAGGAUAUUCCAUUUCCCAGCCC .....(((((((((((.........-----------..((.........))..((((((((((.(((((.....))))).)))))..))))).......)))....))))))))...... ( -35.60) >DroEre_CAF1 8954 120 - 1 AAGCAGGAAACGUUCCCCUCAUUUUUAGCCUCAUUUUAGCACAAUGUAAGCAAUCAGGUGUGCGAGCUGAUUGUCAUUUCCCGCAGUCCUGCAGCCACAGGAUAUUCAAUUUCCCAGCCC ..((.(((((.................((.........)).........((((((((.(.....).))))))))..))))).)).((((((......))))))................. ( -24.70) >consensus AAAAAGGAAAUGGUCCCAUAAUUUU___________UAGCACAAAAUAAGCAAGCAGGUGUGUGAGCUGAUUGUCAGUUGCCGCAGUCCUGCAGCCACAGGAUAUUCCAUUUCCCAGCCC .....(((((((((((......................((.........))..((((((((((.(((((.....))))).)))))..))))).......)))....))))))))...... (-25.30 = -27.17 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:34 2006