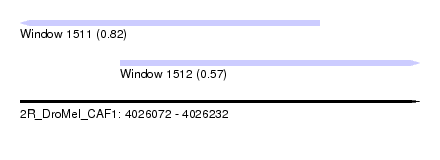
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,026,072 – 4,026,232 |
| Length | 160 |
| Max. P | 0.816936 |

| Location | 4,026,072 – 4,026,192 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.41 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4026072 120 - 20766785 CCGUUCAACGGGGCGUAUGCUCAAUAAUUAAAACGAAUUGGUGUCUACUAAUGAGCAUGCAAGCGGGCACUUUUCUAUUAAUUUAUUUAUGACCCGACAGAGCCAUAAAGGCGGAAACGU (((.....)))((((((((((((.((......((......))......)).)))))))))..(((((((....................)).)))).)...)))......(((....))) ( -32.35) >DroSec_CAF1 8883 120 - 1 CCGUUCAACGGGGCGUAUGCUCAAUAAUUAAAUCGAAUUAGUGUCUCCUAAUGAGCAUGCAAGCGGGCACUUUUCUAUUAAUUUAUUUAUGGCCCGACAGAGCCAUAAAGGCGGAAACGU (((.....)))((((((((((((..............((((......)))))))))))))..((((((.(....................)))))).)...)))......(((....))) ( -34.08) >DroSim_CAF1 10937 120 - 1 CCGUUCAACGGGGCGUAUGCUCAAUAAUUAAAACGAAUUAGUGUCUCCUAAUGAGCAUGCAAGCGGGCACUUUUCUAUUAAUUUAUUUAUGGCCCGACAGAGCCAUAAAGGCGGAAACGU (((.....)))((((((((((((.((......((......))......)).)))))))))..((((((.(....................)))))).)...)))......(((....))) ( -36.05) >DroEre_CAF1 8057 120 - 1 CCGUUCAACGGGGCGUAUGCUCAAUAAUUAAAACGAAUUAGCGUCUCCUAAUGAGCAUGCAGGCGCGCACUUUUCCAUUAAUUUAUUUAUGGCCCAGCAGCGCCAUAAAGGCGGAAACGU (((.....)))...(((((((((.((......(((......)))....)).))))))))).(((((((......((((..........))))....)).)))))......(((....))) ( -35.50) >DroYak_CAF1 10662 119 - 1 -CGUUCAACGGGGCGUAUGCUCAAUAAUUAAAACGAAUUAGCGUCUCCUAAUGAGCAUGCAAGCGCGCACUUUUCUAUUAAUUUAUUUAUGGUCCAACAGAGCCAUAAAGGCGGAAACGU -.((((....))))(((((((((.((......(((......)))....)).))))))))).........................(((((((..(....)..))))))).(((....))) ( -28.90) >consensus CCGUUCAACGGGGCGUAUGCUCAAUAAUUAAAACGAAUUAGUGUCUCCUAAUGAGCAUGCAAGCGGGCACUUUUCUAUUAAUUUAUUUAUGGCCCGACAGAGCCAUAAAGGCGGAAACGU (((.....))).(((((((((((..............((((......)))))))))))))..)).....................((((((((.(....).)))))))).(((....))) (-27.33 = -27.41 + 0.08)

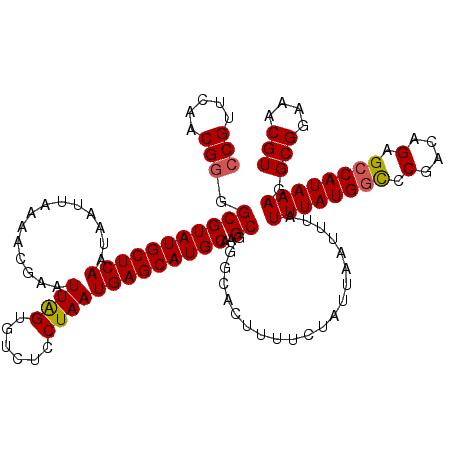
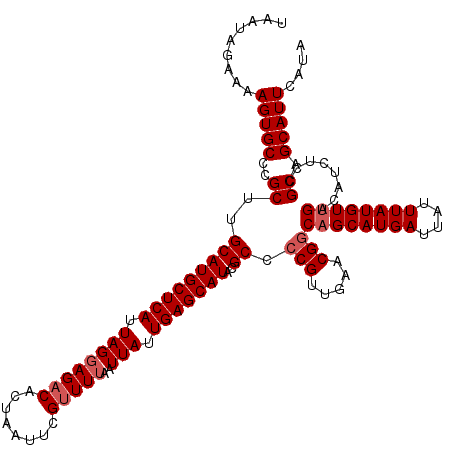
| Location | 4,026,112 – 4,026,232 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -25.42 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4026112 120 + 20766785 UAAUAGAAAAGUGCCCGCUUGCAUGCUCAUUAGUAGACACCAAUUCGUUUUAAUUAUUGAGCAUACGCCCCGUUGAACGGCAGCAUGAUUAUUUAUGUUGACAUCUCCGCAGCAUUCAUA .....(((((((....))))(((((((((.((((((((........))))..)))).)))))))..((.(((.....)))((((((((....))))))))........)).)).)))... ( -29.30) >DroSec_CAF1 8923 120 + 1 UAAUAGAAAAGUGCCCGCUUGCAUGCUCAUUAGGAGACACUAAUUCGAUUUAAUUAUUGAGCAUACGCCCCGUUGAACGGCAGCAUGAUUAUUUAUGUUGACAUCUCCGCAGCAUUCAUA .....(((((((....))))(((((((((...(....)..(((((......))))).)))))))..((.(((.....)))((((((((....))))))))........)).)).)))... ( -27.40) >DroSim_CAF1 10977 120 + 1 UAAUAGAAAAGUGCCCGCUUGCAUGCUCAUUAGGAGACACUAAUUCGUUUUAAUUAUUGAGCAUACGCCCCGUUGAACGGCAGCAUGAUUAUUUAUGUUGACAUCUCCGCAGCAUUCAUA .....(((((((....))))(((((((((.((.(((((........)))))...)).)))))))..((.(((.....)))((((((((....))))))))........)).)).)))... ( -29.40) >DroEre_CAF1 8097 120 + 1 UAAUGGAAAAGUGCGCGCCUGCAUGCUCAUUAGGAGACGCUAAUUCGUUUUAAUUAUUGAGCAUACGCCCCGUUGAACGGCGGCAUGAUUAUUUAUGUGGACAUCUCCGCAGCAUUCAGU .........(((((((....))(((((((.((.((((((......))))))...)).)))))))..((((((.....))).)))...........((((((....))))))))))).... ( -35.60) >DroYak_CAF1 10702 119 + 1 UAAUAGAAAAGUGCGCGCUUGCAUGCUCAUUAGGAGACGCUAAUUCGUUUUAAUUAUUGAGCAUACGCCCCGUUGAACG-CAGCAUGAUUAUUUAUGUUGACAUCUCCGCAGCAUUCAUU .........((((((((...(((((((((.((.((((((......))))))...)).)))))))..))......((...-((((((((....))))))))...))..))).))))).... ( -30.20) >consensus UAAUAGAAAAGUGCCCGCUUGCAUGCUCAUUAGGAGACACUAAUUCGUUUUAAUUAUUGAGCAUACGCCCCGUUGAACGGCAGCAUGAUUAUUUAUGUUGACAUCUCCGCAGCAUUCAUA .........(((((..((..(((((((((.((((((((........)))))..))).)))))))..)).(((.....)))((((((((....))))))))........)).))))).... (-25.42 = -26.22 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:31 2006