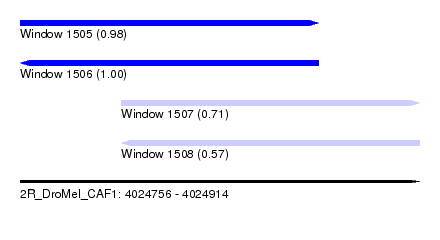
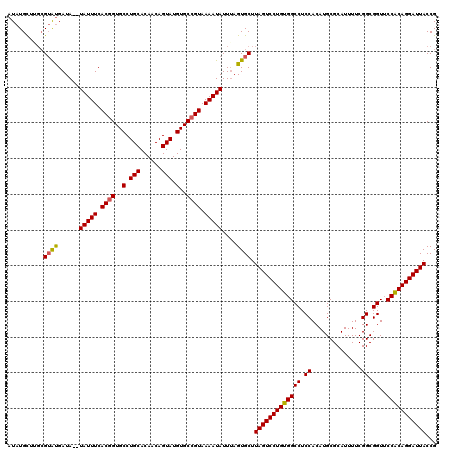
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,024,756 – 4,024,914 |
| Length | 158 |
| Max. P | 0.998586 |

| Location | 4,024,756 – 4,024,874 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -30.86 |
| Energy contribution | -31.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4024756 118 + 20766785 CGGUAAUCCUGUGGAACCGCCGAAAAUGCGCAUGUGGAGGCCACAGGACUAAGCACUUAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUA--UAUCAUUCGCAAGCAUAU .(((..((((((((..((((((......))...))))...))))))))....((.(.(((....))).)(((((......)))))..)))))(((((...--.....)))))........ ( -34.20) >DroSec_CAF1 7569 120 + 1 CGGUAAUCCUGCGGAACCGCCGAAAAUGCGCAUGUGGAGGCCACAGGACUAAGCACUAAAUAUUUUACUGCACAUACUGUUGUGCAGGCACCGUGAAAUAUAUAUCAUACGCAAGCAUAU .((((.(((((.((..((((((......))...))))...)).)))))...........(((((((((((((((......))))))(....)))))))))).))))...(....)..... ( -32.10) >DroSim_CAF1 9621 120 + 1 CGGUAAUCCUGUGGAACCGCCGAAAAUGCGCAUGUGGAGGCCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUAUAUAUCAUACGCAAGCAUAU .((((.((((((((..((((((......))...))))...))))))))...........(((((((((.(((((......))))).(....)))))))))).))))...(....)..... ( -35.70) >DroEre_CAF1 6856 118 + 1 CGGUAAUCCUGUGGCACCGCCGAAAAUGCGCAUGUGGAGGCCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGCUGUGCAGGCACCGUGAAAUA--UAUCGCACGGCAACAUAU (((((.(((((((((.((((((......))...))))..)))))))))............((((((((((......((((...))))...))))))))))--)))))...(....).... ( -40.60) >DroYak_CAF1 9374 118 + 1 CGGUAAUCCUGCGGAACCGCCGAAAAUGCGCAUGUGGAGGCCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUA--UAUCAUACUCCAGCAUAU .((((.(((((.((..((((((......))...))))...)).)))))...........(((((((((.(((((......))))).(....)))))))))--)....))))......... ( -30.60) >consensus CGGUAAUCCUGUGGAACCGCCGAAAAUGCGCAUGUGGAGGCCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUA__UAUCAUACGCAAGCAUAU .((((.((((((((..((((((......))...))))...))))))))............((((((((.(((((......))))).(....)))))))))..)))).............. (-30.86 = -31.26 + 0.40)

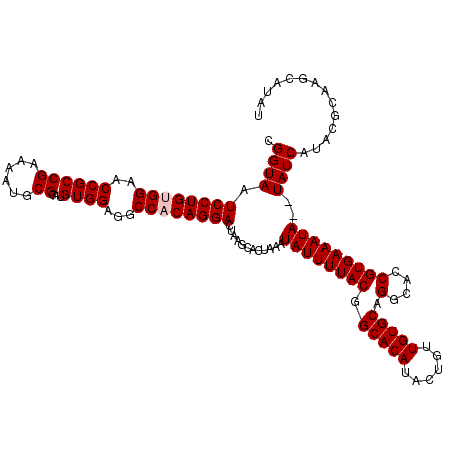
| Location | 4,024,756 – 4,024,874 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4024756 118 - 20766785 AUAUGCUUGCGAAUGAUA--UAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUAAGUGCUUAGUCCUGUGGCCUCCACAUGCGCAUUUUCGGCGGUUCCACAGGAUUACCG ........(((..(((((--(.(((.((((..(.(((......))).)..)))))))))))))..))).(((((((((((((.((..............)).))..)))))))))))... ( -37.44) >DroSec_CAF1 7569 120 - 1 AUAUGCUUGCGUAUGAUAUAUAUUUCACGGUGCCUGCACAACAGUAUGUGCAGUAAAAUAUUUAGUGCUUAGUCCUGUGGCCUCCACAUGCGCAUUUUCGGCGGUUCCGCAGGAUUACCG (((((....))))).............((((..(((((((......))))))).................((((((((((((.((..............)).))..)))))))))))))) ( -34.14) >DroSim_CAF1 9621 120 - 1 AUAUGCUUGCGUAUGAUAUAUAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGGCCUCCACAUGCGCAUUUUCGGCGGUUCCACAGGAUUACCG ..........((((.....((((((.((((..(.(((......))).)..)))).))))))...)))).(((((((((((((.((..............)).))..)))))))))))... ( -37.84) >DroEre_CAF1 6856 118 - 1 AUAUGUUGCCGUGCGAUA--UAUUUCACGGUGCCUGCACAGCAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGGCCUCCACAUGCGCAUUUUCGGCGGUGCCACAGGAUUACCG ..........(..(...(--(((((.((((..(((((...))))...)..)))).))))))...)..).((((((((((((..((....((.(......))))).))))))))))))... ( -42.50) >DroYak_CAF1 9374 118 - 1 AUAUGCUGGAGUAUGAUA--UAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGGCCUCCACAUGCGCAUUUUCGGCGGUUCCGCAGGAUUACCG .......(((((((...(--(((((.((((..(.(((......))).)..)))).))))))...)))))(((((((((((((.((..............)).))..))))))))))))). ( -41.34) >consensus AUAUGCUUGCGUAUGAUA__UAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGGCCUCCACAUGCGCAUUUUCGGCGGUUCCACAGGAUUACCG ..........((((......(((((.((((..(.(((......))).)..)))).)))))....)))).(((((((((((((.((..............)).))..)))))))))))... (-34.50 = -34.34 + -0.16)

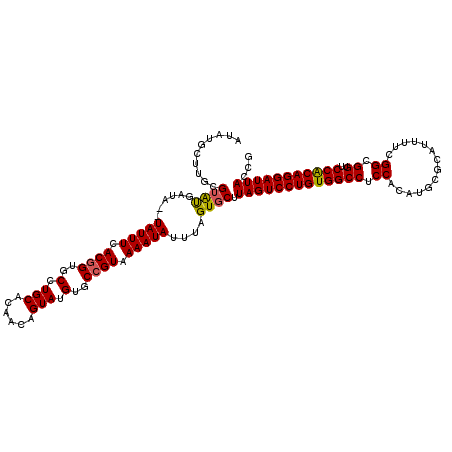
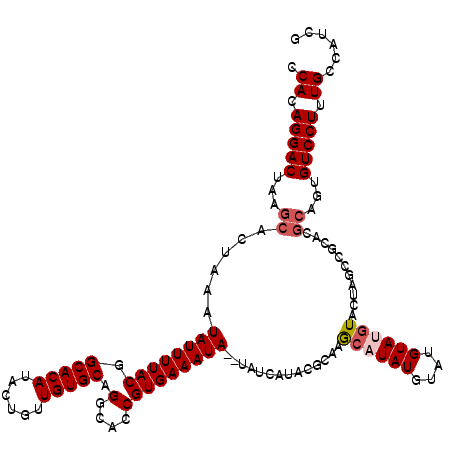
| Location | 4,024,796 – 4,024,914 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.66 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4024796 118 + 20766785 CCACAGGACUAAGCACUUAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUA--UAUCAUUCGCAAGCAUAUGUAUGUACGUACUAGCCGCACGCAGUGUCCUUUGCCAUCG .((.(((((...((.(.(((....))).)))....(((((.((((.(((...(((((...--.....)))))........((((....))))..))))))))))))))))).))...... ( -31.90) >DroSec_CAF1 7609 120 + 1 CCACAGGACUAAGCACUAAAUAUUUUACUGCACAUACUGUUGUGCAGGCACCGUGAAAUAUAUAUCAUACGCAAGCAUAUGUACGUAUGUACUAGCCGCACGCAGUGUCCUUUGCCAUCG .((.(((((...(((.(((.....))).)))....(((((.((((.(((...((((........))))......((((((....))))))....))))))))))))))))).))...... ( -33.10) >DroSim_CAF1 9661 120 + 1 CCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUAUAUAUCAUACGCAAGCAUAUGUACGUAUGUACUAGCCGCACGCAGUGUCCUUUGCCAUCG .((.(((((...((..(((.....)))..))....(((((.((((.(((...((((........))))......((((((....))))))....))))))))))))))))).))...... ( -31.00) >DroEre_CAF1 6896 102 + 1 CCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGCUGUGCAGGCACCGUGAAAUA--UAUCGCACGGCAACAUAUGUAUGUAG-U---------------GGUCCUUUGCCAUCC .((.(((((((.(((....(((((((((((......((((...))))...))))))))))--).......(....).......)))..-)---------------)))))).))...... ( -27.80) >DroYak_CAF1 9414 118 + 1 CCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUA--UAUCAUACUCCAGCAUAUGUAUGUAUGUAGGAGCCGCACGCAGUGUCCUUUGCCAUCG .((.(((((...((.....(((((((((.(((((......))))).(....)))))))))--)......((((.((((((....)))))).))))......))...))))).))...... ( -35.80) >consensus CCACAGGACUAAGCACUAAAUAUUUUACGGCACAUACUGUUGUGCAGGCACCGUGAAAUA__UAUCAUACGCAAGCAUAUGUAUGUAUGUACUAGCCGCACGCAGUGUCCUUUGCCAUCG .((.(((((...((......((((((((.(((((......))))).(....)))))))))..............((((((....))))))...........))...))))).))...... (-22.82 = -23.66 + 0.84)

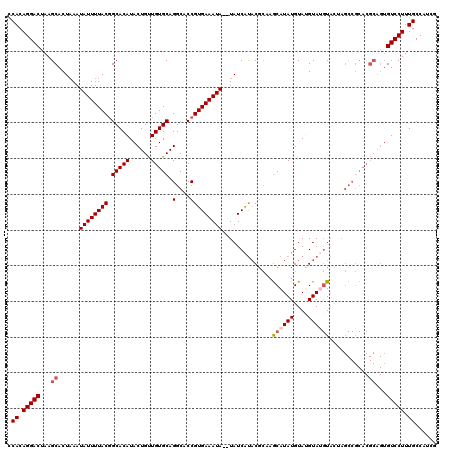
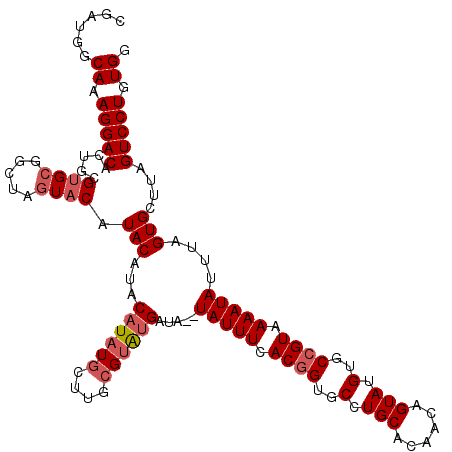
| Location | 4,024,796 – 4,024,914 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -26.15 |
| Energy contribution | -27.34 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4024796 118 - 20766785 CGAUGGCAAAGGACACUGCGUGCGGCUAGUACGUACAUACAUAUGCUUGCGAAUGAUA--UAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUAAGUGCUUAGUCCUGUGG ......((.(((((..(((((((.....))))))).............(((..(((((--(.(((.((((..(.(((......))).)..)))))))))))))..)))...))))).)). ( -35.50) >DroSec_CAF1 7609 120 - 1 CGAUGGCAAAGGACACUGCGUGCGGCUAGUACAUACGUACAUAUGCUUGCGUAUGAUAUAUAUUUCACGGUGCCUGCACAACAGUAUGUGCAGUAAAAUAUUUAGUGCUUAGUCCUGUGG ......((.(((((((((.(((((((..((..(((..((((((((....)))))).))..)))...))...))).))))..))))....(((.(((.....))).)))...))))).)). ( -34.90) >DroSim_CAF1 9661 120 - 1 CGAUGGCAAAGGACACUGCGUGCGGCUAGUACAUACGUACAUAUGCUUGCGUAUGAUAUAUAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGG ......((.(((((...(((.(((....((((....))))...))).)))((((.....((((((.((((..(.(((......))).)..)))).))))))...))))...))))).)). ( -37.10) >DroEre_CAF1 6896 102 - 1 GGAUGGCAAAGGACC---------------A-CUACAUACAUAUGUUGCCGUGCGAUA--UAUUUCACGGUGCCUGCACAGCAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGG (((((((((((....---------------.-)).(((....))))))))(..(...(--(((((.((((..(((((...))))...)..)))).))))))...)..)...))))..... ( -30.70) >DroYak_CAF1 9414 118 - 1 CGAUGGCAAAGGACACUGCGUGCGGCUCCUACAUACAUACAUAUGCUGGAGUAUGAUA--UAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGG ......((.(((((...((.....(((((..((((......))))..)))))((...(--(((((.((((..(.(((......))).)..)))).))))))...))))...))))).)). ( -36.00) >consensus CGAUGGCAAAGGACACUGCGUGCGGCUAGUACAUACAUACAUAUGCUUGCGUAUGAUA__UAUUUCACGGUGCCUGCACAACAGUAUGUGCCGUAAAAUAUUUAGUGCUUAGUCCUGUGG ......((.(((((.....((((.....)))).(((...((((((....)))))).....(((((.((((..(.(((......))).)..)))).)))))....)))....))))).)). (-26.15 = -27.34 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:27 2006