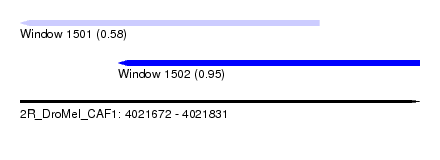
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,021,672 – 4,021,831 |
| Length | 159 |
| Max. P | 0.949762 |

| Location | 4,021,672 – 4,021,791 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -28.68 |
| Energy contribution | -29.84 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
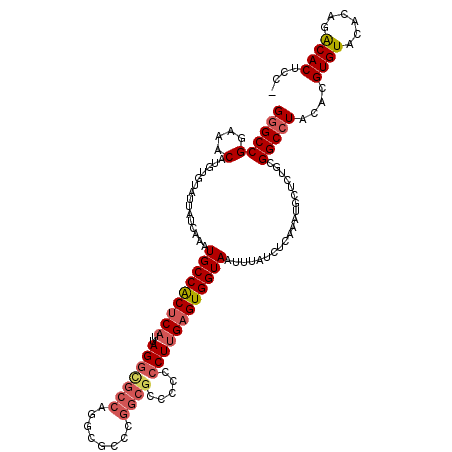
>2R_DroMel_CAF1 4021672 119 - 20766785 GGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGAUGCCAGGCGCCUGGCGCCCCCCCUUGAGUGGUAAUUUAUCUCAAAUGCUCGGCGGCCUACACGUGUACACAGACACUCC- ((((((....).((.(((((((.((.(((((((((.....((((((....))))))........))))))))).)).))....))))).))..)))))....((((......))))...- ( -38.82) >DroSec_CAF1 4469 119 - 1 GGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCAGGCACCCGGCGCUCCCCCUUGAGUGGUAAUUUAUCUCAAAUGCUCUGCGGCCUACACGUGUACACAGACACUGC- ((((((....)..(.(((((((.((.(((((((((...((((((.(....).))))))......))))))))).)).))....))))).)...)))))....((((......))))...- ( -40.30) >DroSim_CAF1 6525 119 - 1 GGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCAGGCGCCCGGCGCUCCCCCUUGAGUGGUAAUUUAUCUCAAAUGCUCUGCGGCCUACACGUGUACACAGACACUGC- ((((((....)..(.(((((((.((.(((((((((...((((((.(....).))))))......))))))))).)).))....))))).)...)))))....((((......))))...- ( -40.30) >DroEre_CAF1 3943 113 - 1 GGGCCGGAAACAUGUGUAUUAUCAAAUGCCGCUCAUUAGGCUCCCUGCCCCC------ACGCCUGGAGUGGUAAUUUAUCUCAAAUGCUCUGCGGCCUACACGUGUACACAGACACUCC- ((((((....)..(.(((((((.((.((((((((..(((((...........------..))))))))))))).)).))....))))).)...)))))....((((......))))...- ( -32.02) >DroYak_CAF1 6104 119 - 1 G-GCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCCGUCGCCCGGCGGCCCCCCUUGUGUGGUAAUUUAUCUCAAACGCUCUGCGGCCUACACGUGUACAUGGGCACUCCA (-.(((...(((((((((........((((((.((...((.(.(((((....))))).)))...)).))))))............(((...)))...)))))))))...))).)...... ( -39.20) >consensus GGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCAGGCGCCCGGCGCCCCCCCUUGAGUGGUAAUUUAUCUCAAAUGCUCUGCGGCCUACACGUGUACACAGACACUCC_ ((((((....)...............(((((((((..(((((((........)))).....))))))))))))....................)))))....((((......)))).... (-28.68 = -29.84 + 1.16)

| Location | 4,021,711 – 4,021,831 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -42.84 |
| Consensus MFE | -33.58 |
| Energy contribution | -34.46 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
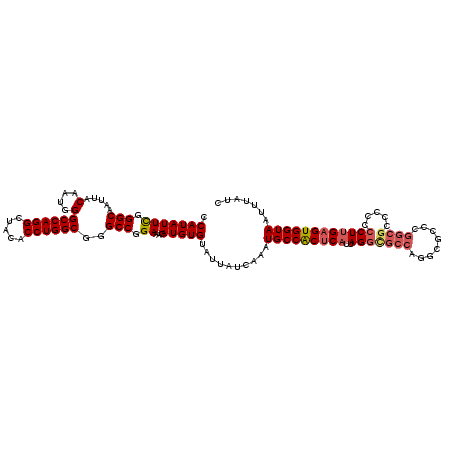
>2R_DroMel_CAF1 4021711 120 - 20766785 CCAUAUUUGGGCAAUUACACUGGGCCAGGCUACACCUGGCGGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGAUGCCAGGCGCCUGGCGCCCCCCCUUGAGUGGUAAUUUAUC .....(((((.....(((((..((((..((((....)))).))))(....)..)))))...)))))(((((((((.....((((((....))))))........)))))))))....... ( -44.02) >DroSec_CAF1 4508 120 - 1 CCAUAUUCGGGCAAUUACAAUGGGCCAGGCUACACCUGGCGGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCAGGCACCCGGCGCUCCCCCUUGAGUGGUAAUUUAUC .(((((((.(((.....(....)((((((.....))))))..))).))...)))))..........(((((((((...((((((.(....).))))))......)))))))))....... ( -44.10) >DroSim_CAF1 6564 120 - 1 CCAUAUUCGGGCAAUUACAAUGGGCCAGGCUACACCUGGCGGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCAGGCGCCCGGCGCUCCCCCUUGAGUGGUAAUUUAUC .(((((((.(((.....(....)((((((.....))))))..))).))...)))))..........(((((((((...((((((.(....).))))))......)))))))))....... ( -44.10) >DroEre_CAF1 3982 114 - 1 CCAUAUUCGGGCAAUUACAAUGGGCCAGGCUACACCUGGCGGGCCGGAAACAUGUGUAUUAUCAAAUGCCGCUCAUUAGGCUCCCUGCCCCC------ACGCCUGGAGUGGUAAUUUAUC ((((.(((((((...........((((((.....))))))((((.((..((....))..........(((........)))..)).))))..------..)))))))))))......... ( -40.20) >DroYak_CAF1 6144 119 - 1 CCAUAUUUGGGCAAUUACAAUGGGCCAGGCUACACCUGGCG-GCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCCGUCGCCCGGCGGCCCCCCUUGUGUGGUAAUUUAUC ((((((..(((............((((((.....))))))(-((((....)................(((........)))(((........))))))).)))..))))))......... ( -41.80) >consensus CCAUAUUCGGGCAAUUACAAUGGGCCAGGCUACACCUGGCGGGCCGGAAACAUGUGUAUUAUCAAAUGCCACUCAUUAGGCGCCAGGCGCCCGGCGCCCCCCCUUGAGUGGUAAUUUAUC .(((((((.(((.....(....)((((((.....))))))..))).))...)))))..........(((((((((..(((((((........)))).....))))))))))))....... (-33.58 = -34.46 + 0.88)

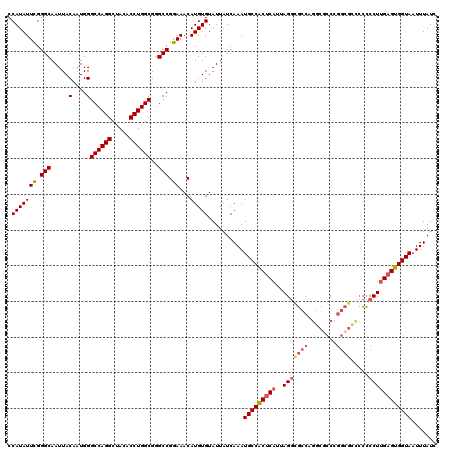
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:21 2006