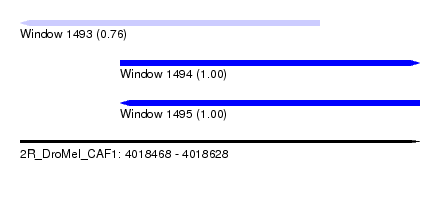
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,018,468 – 4,018,628 |
| Length | 160 |
| Max. P | 0.998600 |

| Location | 4,018,468 – 4,018,588 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -28.25 |
| Energy contribution | -30.06 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4018468 120 - 20766785 GGCAAGCCGAUUAGGGCUUGGGUCCGUCAAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGAAAAUGGCCUAUAAAUUUUACAUUUAACGCGGCCCGAGGAUGACUCUUUUC ((((((((......))))))...))((((..(((..((((((.....)))))).(((((..(((((.......)))))..(((((.....))))).)))))...)))..))))....... ( -37.40) >DroSec_CAF1 1323 119 - 1 GGCAAGCCGAUUAGGGCUCGGGUCCGCCCAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG-AAUGGCCUAUAAAUUUUACAUUUAACGCGGCCCGAGGAUGACUCUUUUC ((....))((..((((((((((.((((.........((((((.....))))))........(((((....-..)))))...................)))))))))).....))))..)) ( -39.30) >DroSim_CAF1 3375 119 - 1 GGCAAGCCGAUUAGGGCUCGGGUCCGCCCAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCGACGG-AAUGGCCUAUAAAUUUUACAUUUAACGCGGCCCGAGGAUGACUCUUUUC ((....))((..((((((((((.((((.........((((((.....))))))........(((((....-..)))))...................)))))))))).....))))..)) ( -38.10) >DroEre_CAF1 1316 104 - 1 GGCAAGCCGAUUAG-GC-CGGGUCCGUCCAGCUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG-AAUGGCCUAUAAAUUUUACAUUUC-------------AUGACUCUUUUC ((....))...(((-((-((..((((((((((....((((((.....)))))).....)))))...))))-).)))))))...............-------------............ ( -36.10) >DroYak_CAF1 3210 118 - 1 GGCAAGCCGAUUAGGGC-CGGGUCCGUCAGACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG-AAUGGCCUAUAAAUUUUACAUUUAACGCGGCCCGAGGAUGACUCUUUUC ((....)).....((((-((((((.....)))....((((((.....)))))).))(((..(((((....-..)))))..(((((.....))))).))))))))(((.....)))..... ( -37.60) >consensus GGCAAGCCGAUUAGGGCUCGGGUCCGUCCAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG_AAUGGCCUAUAAAUUUUACAUUUAACGCGGCCCGAGGAUGACUCUUUUC ((....))((..((((((((((.((((.........((((((.....))))))........(((((.......)))))...................)))))))))).....))))..)) (-28.25 = -30.06 + 1.81)

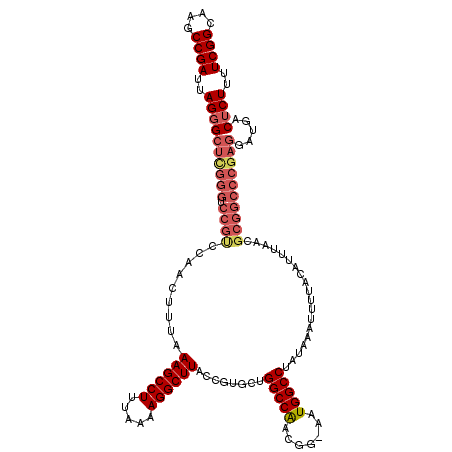
| Location | 4,018,508 – 4,018,628 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -33.78 |
| Energy contribution | -34.46 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4018508 120 + 20766785 UAGGCCAUUUUCGUUGGCCAGCACGGUAAGCCUUUUAAAGGCUUUAAAGUUUGACGGACCCAAGCCCUAAUCGGCUUGCCAUGCUAAACACGCUUUUUUGUCUACGGAAAAUUCAAUGUA ..(((((.......)))))((((.((((((((..(((..(((((....(((.....)))..))))).)))..)))))))).))))........(((((((....)))))))......... ( -37.70) >DroSec_CAF1 1363 119 + 1 UAGGCCAUU-CCGUUGGCCAGCACGGUAAGCCUUUUAAAGGCUUUAAAGUUGGGCGGACCCGAGCCCUAAUCGGCUUGCCAUGCUAAACACGCUUUUUUGUCUACGGAAAAUUCAAUGUA ..(((((..-....)))))((((.((((((((..(((..(((((.......(((....)))))))).)))..)))))))).))))........(((((((....)))))))......... ( -40.81) >DroSim_CAF1 3415 119 + 1 UAGGCCAUU-CCGUCGGCCAGCACGGUAAGCCUUUUAAAGGCUUUAAAGUUGGGCGGACCCGAGCCCUAAUCGGCUUGCCAUGCUAAACACGCUUUUUUGUCUACGGAAAAUUCAAUGUA ..((((...-.....))))((((.((((((((..(((..(((((.......(((....)))))))).)))..)))))))).))))........(((((((....)))))))......... ( -38.91) >DroEre_CAF1 1343 117 + 1 UAGGCCAUU-CCGUUGGCCAGCACGGUAAGCCUUUUAAAGGCUUUAAAGCUGGACGGACCCG-GC-CUAAUCGGCUUGCCAUGCUAAACACGCUUUUUUGUCUACGGAAAAUCCAAUGUA ..(((((..-....)))))((((.((((((((..(((..((((....))))((.((....))-.)-))))..)))))))).))))..(((.(.(((((((....))))))).)...))). ( -39.30) >DroYak_CAF1 3250 118 + 1 UAGGCCAUU-CCGUUGGCCAGCACGGUAAGCCUUUUAAAGGCUUUAAAGUCUGACGGACCCG-GCCCUAAUCGGCUUGCCAUGCUAAACACGCUUUUUUGUCUACGGAAAAUUCAAUGUA ..(((((..-....)))))((((.((((((((..(((..((((.....(((.....)))..)-))).)))..)))))))).))))........(((((((....)))))))......... ( -38.90) >consensus UAGGCCAUU_CCGUUGGCCAGCACGGUAAGCCUUUUAAAGGCUUUAAAGUUGGACGGACCCGAGCCCUAAUCGGCUUGCCAUGCUAAACACGCUUUUUUGUCUACGGAAAAUUCAAUGUA ..(((((.......)))))((((.((((((((..(((..(((((....(((.....)))..))))).)))..)))))))).))))........(((((((....)))))))......... (-33.78 = -34.46 + 0.68)

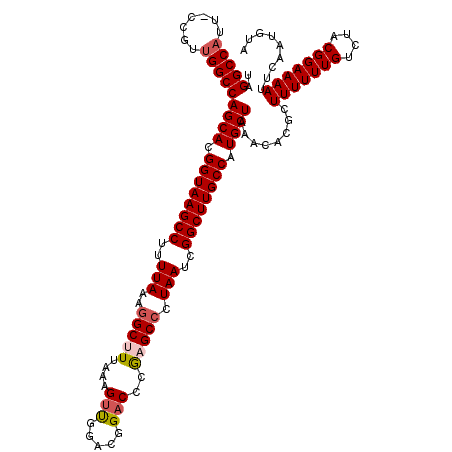
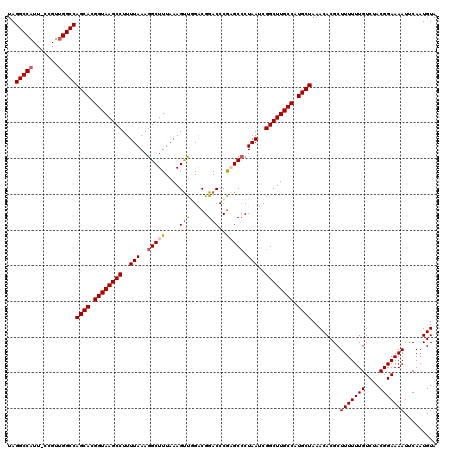
| Location | 4,018,508 – 4,018,628 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -34.58 |
| Energy contribution | -34.86 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4018508 120 - 20766785 UACAUUGAAUUUUCCGUAGACAAAAAAGCGUGUUUAGCAUGGCAAGCCGAUUAGGGCUUGGGUCCGUCAAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGAAAAUGGCCUA ................((((((........))))))((((((.(((((....(((((((.(((.......)))...))))))).....))))).)))))).(((((.......))))).. ( -38.20) >DroSec_CAF1 1363 119 - 1 UACAUUGAAUUUUCCGUAGACAAAAAAGCGUGUUUAGCAUGGCAAGCCGAUUAGGGCUCGGGUCCGCCCAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG-AAUGGCCUA ................((((((........))))))((((((.(((((....((((((.(((....)))........)))))).....))))).)))))).(((((....-..))))).. ( -39.30) >DroSim_CAF1 3415 119 - 1 UACAUUGAAUUUUCCGUAGACAAAAAAGCGUGUUUAGCAUGGCAAGCCGAUUAGGGCUCGGGUCCGCCCAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCGACGG-AAUGGCCUA ...........((((((.((((........))))((((((((.(((((....((((((.(((....)))........)))))).....))))).))))))))....))))-))....... ( -38.30) >DroEre_CAF1 1343 117 - 1 UACAUUGGAUUUUCCGUAGACAAAAAAGCGUGUUUAGCAUGGCAAGCCGAUUAG-GC-CGGGUCCGUCCAGCUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG-AAUGGCCUA ...(((((.(((.(((((((((........)))))...)))).))))))))(((-((-((..((((((((((....((((((.....)))))).....)))))...))))-).))))))) ( -42.60) >DroYak_CAF1 3250 118 - 1 UACAUUGAAUUUUCCGUAGACAAAAAAGCGUGUUUAGCAUGGCAAGCCGAUUAGGGC-CGGGUCCGUCAGACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG-AAUGGCCUA ................((((((........))))))((((((.(((((....(((((-.(((((.....)))))....))))).....))))).)))))).(((((....-..))))).. ( -38.70) >consensus UACAUUGAAUUUUCCGUAGACAAAAAAGCGUGUUUAGCAUGGCAAGCCGAUUAGGGCUCGGGUCCGUCCAACUUUAAAGCCUUUAAAAGGCUUACCGUGCUGGCCAACGG_AAUGGCCUA ................((((((........))))))((((((.(((((....((((((.((((.......))))...)))))).....))))).)))))).(((((.......))))).. (-34.58 = -34.86 + 0.28)

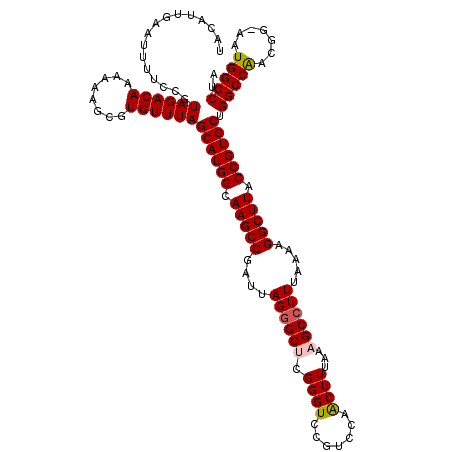
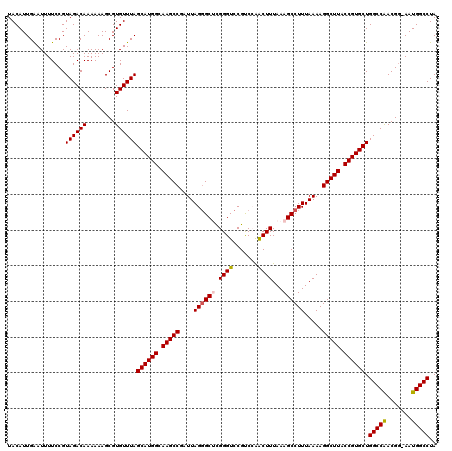
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:14 2006